bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-10_CDS_annotation_glimmer3.pl_2_5
Length=674
Score E
Sequences producing significant alignments: (Bits) Value
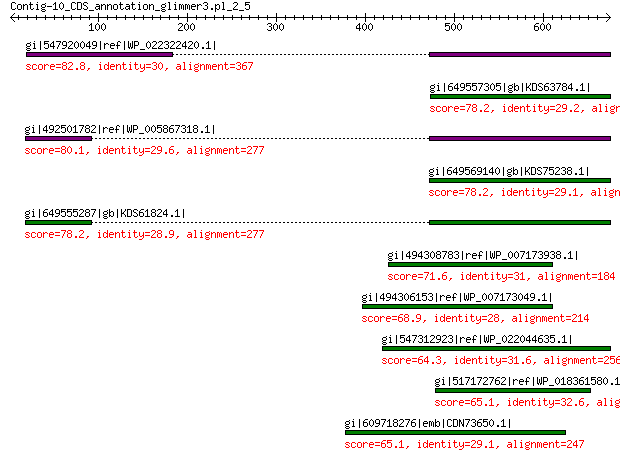
gi|547920049|ref|WP_022322420.1| capsid protein VP1 82.8 3e-13
gi|649557305|gb|KDS63784.1| capsid family protein 78.2 1e-12
gi|492501782|ref|WP_005867318.1| hypothetical protein 80.1 2e-12
gi|649569140|gb|KDS75238.1| capsid family protein 78.2 4e-12
gi|649555287|gb|KDS61824.1| capsid family protein 78.2 7e-12
gi|494308783|ref|WP_007173938.1| hypothetical protein 71.6 9e-10
gi|494306153|ref|WP_007173049.1| hypothetical protein 68.9 5e-09
gi|547312923|ref|WP_022044635.1| putative uncharacterized protein 64.3 9e-08
gi|517172762|ref|WP_018361580.1| hypothetical protein 65.1 9e-08
gi|609718276|emb|CDN73650.1| conserved hypothetical protein 65.1 1e-07
>gi|547920049|ref|WP_022322420.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
gi|524592961|emb|CDD13573.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
Length=553
Score = 82.8 bits (203), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 64/207 (31%), Positives = 97/207 (47%), Gaps = 9/207 (4%)
Query 472 HIETPIYLGGSSLEIEFQEVVNNSGT-EDQPLGTLAGRGVATNHKGGNIVFKA--DEPGY 528
++ P +LGG + I EV+ S T E P +AG G++ G N FK +E GY
Sbjct 352 RLQRPQFLGGGRMPISVSEVLQTSSTDETSPQANMAGHGISA---GINNGFKHYFEEHGY 408
Query 529 LFCITSITPRVDYFQGNEWDMYLESLDDLHKPQLDGIGFQDRLYKHINANTDSTEFNKTV 588
+ I SITPR Y QG D D + P+ + Q+ + + + D+ N T
Sbjct 409 IIGIMSITPRSGYQQGVPRDFTKFDNMDFYFPEFAHLSEQEIKNQELFVSEDAAYNNGTF 468
Query 589 GKQPAWIEYMTNVNKTYGNFALVENEGWMCLNRIFGD-INTYTTYIFPHLYNNIFADTDV 647
G P + EY + ++ +G+F N + LNRIF D N TT++ N +FA ++
Sbjct 469 GYTPRYAEYKYHPSEAHGDFR--GNLSFWHLNRIFEDKPNLNTTFVECKPSNRVFATSET 526
Query 648 TSQNFWVQIAFNTNPRRVMSAKVIPNL 674
FWVQ+ + R+M P L
Sbjct 527 EDDKFWVQMYQDVKALRLMPKYGTPML 553
Score = 50.4 bits (119), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 46/177 (26%), Positives = 77/177 (44%), Gaps = 19/177 (11%)
Query 19 RLNNYNRSTHDLSFVMRTSMAPGVLVPTLKMLMLPGDTFPVKTRCHTLTHPTVGPLFGSF 78
R+ R+ +LS+ + ++ G LVP + M ++ GD F VKT P V P+
Sbjct 9 RMKRPRRNAFNLSYESKLTLNMGELVPIMCMPVVSGDKFRVKTESLVRLAPLVAPMMHRV 68
Query 79 KQQNDFFFCPIRL-YNAMLHNNALNIGLDMKKVKLPIVRIIASDLDLTKKMKGSNGTLKK 137
+FF P RL +N + + G+D + +P+ I + D + S +K+
Sbjct 69 NVFTHYFFVPNRLVWNEW--EDFITKGVDGE--DMPMFPKIQINQD--SHLVSSASLIKE 122
Query 138 MIHPSSLVKTLGLSNLEKNNSQQWD------------WNAIPILAYFDIFKNYYANK 182
SSL LGL L ++ +D +A+P AY I+ YY ++
Sbjct 123 YFGDSSLWDYLGLPTLSACGNKSYDVVNGVKVPSGFQVSALPFRAYQLIYNEYYRDQ 179
>gi|649557305|gb|KDS63784.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649559156|gb|KDS65543.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=245
Score = 78.2 bits (191), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 59/204 (29%), Positives = 92/204 (45%), Gaps = 5/204 (2%)
Query 473 IETPIYLGGSSLEIEFQEVVNNSGTED-QPLGTLAGRGVATNHKGGNIVFKADEPGYLFC 531
++ P +LGG I EV+ S T+ P +AG G++ G + +E GY+
Sbjct 45 LQRPQFLGGGRTPISVSEVLQTSSTDSTSPQANMAGHGISAGVNHGFTRY-FEEHGYIMG 103
Query 532 ITSITPRVDYFQGNEWDMYLESLDDLHKPQLDGIGFQDRLYKHINANTDSTEFNKTVGKQ 591
I SI PR Y QG D D + P+ +G Q+ + + N T G
Sbjct 104 IMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEELYLNESDAANEGTFGYT 163
Query 592 PAWIEYMTNVNKTYGNFALVENEGWMCLNRIFGDI-NTYTTYIFPHLYNNIFADTDVTSQ 650
P + EY + N+ +G+F N + LNRIF + N TT++ + N +FA + +
Sbjct 164 PRYAEYKYSQNEVHGDFR--GNMAFWHLNRIFKEKPNLNTTFVECNPSNRVFATAETSDD 221
Query 651 NFWVQIAFNTNPRRVMSAKVIPNL 674
+WVQI + R+M P L
Sbjct 222 KYWVQIYQDIKALRLMPKYGTPML 245
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 80.1 bits (196), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 60/207 (29%), Positives = 95/207 (46%), Gaps = 9/207 (4%)
Query 472 HIETPIYLGGSSLEIEFQEVVNNSGTED-QPLGTLAGRGVATNHKGGNIVFKA--DEPGY 528
++ P +LGG I EV+ S T+ P +AG G++ G N FK +E GY
Sbjct 337 RLQRPQFLGGGRTPISVSEVLQTSATDSTSPQANMAGHGISA---GVNHGFKRYFEEHGY 393
Query 529 LFCITSITPRVDYFQGNEWDMYLESLDDLHKPQLDGIGFQDRLYKHINANTDSTEFNKTV 588
+ I SI PR Y QG D D + P+ +G Q+ + + N T
Sbjct 394 IIGIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEEVYLQQTPASNNGTF 453
Query 589 GKQPAWIEYMTNVNKTYGNFALVENEGWMCLNRIFGD-INTYTTYIFPHLYNNIFADTDV 647
G P + EY ++N+ +G+F N + LNRIF + N TT++ + N +FA +
Sbjct 454 GYTPRYAEYKYSMNEVHGDFR--GNMAFWHLNRIFSESPNLNTTFVECNPSNRVFATAET 511
Query 648 TSQNFWVQIAFNTNPRRVMSAKVIPNL 674
+ +W+Q+ + R+M P L
Sbjct 512 SDDKYWIQLYQDVKALRLMPKYGTPML 538
Score = 39.7 bits (91), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 22/74 (30%), Positives = 33/74 (45%), Gaps = 0/74 (0%)
Query 18 TRLNNYNRSTHDLSFVMRTSMAPGVLVPTLKMLMLPGDTFPVKTRCHTLTHPTVGPLFGS 77
+L R+ +LS+ + + G LVP + ++PGD F V T P V P+
Sbjct 8 VKLKRPRRNVFNLSYENKLTANAGELVPIMCKPVVPGDKFRVNTEMLVRLAPLVAPMMHR 67
Query 78 FKQQNDFFFCPIRL 91
+FF P RL
Sbjct 68 VDVFTHYFFVPNRL 81
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 78.2 bits (191), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 59/205 (29%), Positives = 92/205 (45%), Gaps = 5/205 (2%)
Query 472 HIETPIYLGGSSLEIEFQEVVNNSGTED-QPLGTLAGRGVATNHKGGNIVFKADEPGYLF 530
++ P +LGG I EV+ S T+ P +AG G++ G + +E GY+
Sbjct 189 RLQRPQFLGGGRTPISVSEVLQTSSTDSTSPQANMAGHGISAGVNHGFTRY-FEEHGYIM 247
Query 531 CITSITPRVDYFQGNEWDMYLESLDDLHKPQLDGIGFQDRLYKHINANTDSTEFNKTVGK 590
I SI PR Y QG D D + P+ +G Q+ + + N T G
Sbjct 248 GIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEELYLNESDAANEGTFGY 307
Query 591 QPAWIEYMTNVNKTYGNFALVENEGWMCLNRIFGD-INTYTTYIFPHLYNNIFADTDVTS 649
P + EY + N+ +G+F N + LNRIF + N TT++ + N +FA + +
Sbjct 308 TPRYAEYKYSQNEVHGDFR--GNMAFWHLNRIFKEKPNLNTTFVECNPSNRVFATAETSD 365
Query 650 QNFWVQIAFNTNPRRVMSAKVIPNL 674
+WVQI + R+M P L
Sbjct 366 DKYWVQIYQDIKALRLMPKYGTPML 390
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 78.2 bits (191), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 59/205 (29%), Positives = 92/205 (45%), Gaps = 5/205 (2%)
Query 472 HIETPIYLGGSSLEIEFQEVVNNSGTED-QPLGTLAGRGVATNHKGGNIVFKADEPGYLF 530
++ P +LGG I EV+ S T+ P +AG G++ G + +E GY+
Sbjct 340 RLQRPQFLGGGRTPISVSEVLQTSSTDSTSPQANMAGHGISAGVNHGFTRY-FEEHGYIM 398
Query 531 CITSITPRVDYFQGNEWDMYLESLDDLHKPQLDGIGFQDRLYKHINANTDSTEFNKTVGK 590
I SI PR Y QG D D + P+ +G Q+ + + N T G
Sbjct 399 GIMSIRPRTGYQQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEELYLNESDAANEGTFGY 458
Query 591 QPAWIEYMTNVNKTYGNFALVENEGWMCLNRIFGD-INTYTTYIFPHLYNNIFADTDVTS 649
P + EY + N+ +G+F N + LNRIF + N TT++ + N +FA + +
Sbjct 459 TPRYAEYKYSQNEVHGDFR--GNMAFWHLNRIFKEKPNLNTTFVECNPSNRVFATAETSD 516
Query 650 QNFWVQIAFNTNPRRVMSAKVIPNL 674
+WVQI + R+M P L
Sbjct 517 DKYWVQIYQDIKALRLMPKYGTPML 541
Score = 40.0 bits (92), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 34/74 (46%), Gaps = 0/74 (0%)
Query 18 TRLNNYNRSTHDLSFVMRTSMAPGVLVPTLKMLMLPGDTFPVKTRCHTLTHPTVGPLFGS 77
+L R+ +LS+ + ++ G L+P + ++PGD F V T P V P+
Sbjct 8 VKLKRPRRNVFNLSYENKLTVNAGELIPIMCKPVVPGDKFRVNTEMLVRLAPLVAPMMHR 67
Query 78 FKQQNDFFFCPIRL 91
+FF P RL
Sbjct 68 VDVFTHYFFVPNRL 81
>gi|494308783|ref|WP_007173938.1| hypothetical protein [Prevotella bergensis]
gi|270333035|gb|EFA43821.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=553
Score = 71.6 bits (174), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 57/200 (29%), Positives = 94/200 (47%), Gaps = 25/200 (13%)
Query 426 DTSGGSFTLDTLNLAKKVYTMLNRIAISDGSYNAWIQTVYTSGGLNHIETPI-------Y 478
D+S G F++ +L A V +L+ + ++ ++ Y +E P Y
Sbjct 290 DSSEGDFSVSSLRAAFAVDKLLSVTMRAGKTFQDQMRAHYG------VEIPDSRDGRVNY 343
Query 479 LGGSSLEIEFQEVVNNSGT---EDQP----LGTLAGRGVATNHKGGNIVFKADEPGYLFC 531
LGG +++ +V SGT E +P LG +AG+G + G IVF A E G L C
Sbjct 344 LGGFDSDMQVSDVTQTSGTTATEYKPEAGYLGRVAGKGTGSGR--GRIVFDAKEHGVLMC 401
Query 532 ITSITPRVDYFQGNEWDMYLESLD--DLHKPQLDGIGFQDRLYKHINANTDSTEFNKTVG 589
I S+ P++ Y D ++ LD D P+ + +G Q +I++ + N +G
Sbjct 402 IYSLVPQIQY-DCTRLDPMVDKLDRFDYFTPEFENLGMQPLNSSYISSFCTTDPKNPVLG 460
Query 590 KQPAWIEYMTNVNKTYGNFA 609
QP + EY T ++ +G FA
Sbjct 461 YQPRYSEYKTALDVNHGQFA 480
>gi|494306153|ref|WP_007173049.1| hypothetical protein [Prevotella bergensis]
gi|270333881|gb|EFA44667.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=519
Score = 68.9 bits (167), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 60/234 (26%), Positives = 104/234 (44%), Gaps = 29/234 (12%)
Query 396 LKTYQSDVNTNWVNTEWLDGDSG----INSITAIDTSGGSFTLDTLNLAKKVYTMLNRIA 451
+ + D + N+ ++ D +N +D + G F++ +L A V +L+
Sbjct 222 IPEFSDDEHLNFDRDQYADQSKSNFTQLNFPVDVDNNLGYFSVSSLRSAFAVDKLLSVTM 281
Query 452 ISDGSYNAWIQTVYTSGGLNHIETPI-------YLGGSSLEIEFQEVVNNSGT---EDQP 501
+ ++ ++ Y +E P YLGG +++ +V SGT E +P
Sbjct 282 RAGKTFQDQMRAHYG------VEIPDSRDGRVNYLGGFDSDLQVSDVTQTSGTTATEYKP 335
Query 502 ----LGTLAGRGVATNHKGGNIVFKADEPGYLFCITSITPRVDYFQGNEWDMYLESLD-- 555
LG +AG+G + G IVF A E G L CI S+ P++ Y D ++ LD
Sbjct 336 EAGYLGRIAGKGTGSGR--GRIVFDAKEHGVLMCIYSLVPQIQY-DCTRLDPMVDKLDRF 392
Query 556 DLHKPQLDGIGFQDRLYKHINANTDSTEFNKTVGKQPAWIEYMTNVNKTYGNFA 609
D P+ + +G Q +I++ N +G QP + EY T ++ +G FA
Sbjct 393 DFFTPEFENLGMQPLNSSYISSFCTPDPKNPVLGYQPRYSEYKTALDINHGQFA 446
>gi|547312923|ref|WP_022044635.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
gi|524208404|emb|CCZ76639.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
Length=338
Score = 64.3 bits (155), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 81/303 (27%), Positives = 126/303 (42%), Gaps = 49/303 (16%)
Query 419 INSITAID---TSGGSFTLDTLNLAKKVYTMLNRIAISDGSYNAWIQTVY-TSGGLNHIE 474
I + A+D ++G S + L L K+ ++R+ +S G +T++ T ++
Sbjct 36 IEVMNALDLNISTGFSVAVPELRLRTKIQNWMDRLFVSGGRVGDVFRTLWGTKSSAIYVN 95
Query 475 TPIYLGGSSLEIE---FQEVVNNSGT-EDQPLGTLAG----RGVATNHKGGNIVFKADEP 526
P +LG I + + N S + ED LG LA + H G I + A EP
Sbjct 96 KPDFLGVWQASINPSNVRAMANGSASGEDANLGQLAACVDRYCDFSGHSG--IDYYAKEP 153
Query 527 GYLFCITSITPRVDYFQGNEWDMYLESLDDLHKPQLDGIGFQ----------DRLYKHIN 576
G IT + P Y QG D+ S D P+L+GIGFQ R +
Sbjct 154 GTFMLITMLVPEPAYSQGLHPDLASISFGDDFNPELNGIGFQLVPRHRFSMMPRGFNFTG 213
Query 577 ANTDSTE-FNKT------------VGKQPAWIEYMTNVNKTYGNFALVENEGWMCLNR-- 621
+ +++ F T VG++ AW T+ ++ +G+FA N + L R
Sbjct 214 LDQEASPWFGHTGTGVLVDPNMVSVGEEVAWSWLRTDYSRLHGDFAQNGNYQYWVLTRRF 273
Query 622 --IFGDINT-------YT-TYIFPHLYNNIFADTDVTSQNFWVQIAFNTNPRRVMSAKVI 671
F D T YT TYI P + +F D + + NF F+ N +SA +
Sbjct 274 TTYFPDDGTGFYQDGEYTGTYINPLDWQYVFVDQTLMAGNFAYYGTFDLNVTSSLSANYM 333
Query 672 PNL 674
P L
Sbjct 334 PYL 336
>gi|517172762|ref|WP_018361580.1| hypothetical protein [Prevotella nanceiensis]
Length=568
Score = 65.1 bits (157), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 57/198 (29%), Positives = 89/198 (45%), Gaps = 24/198 (12%)
Query 478 YLGGSSLEIEFQEVVNNSGT-----EDQPLGTLAGR--GVATNHKGGNIVFKADEPGYLF 530
Y+GG I+ +V +SGT +D G GR G AT G+I F A E G L
Sbjct 351 YIGGFDSNIQVGDVTQSSGTTVTGTKDTSFGGYLGRTTGKATGSGSGHIRFDAKEHGILM 410
Query 531 CITSITPRVDYFQGNEWDMYLESLD--DLHKPQLDGIGFQDRLYKHINANTDSTEFNKTV 588
CI S+ P V Y D +++ ++ D P+ + +G Q K+I+ ++ N +
Sbjct 411 CIYSLVPDVQY-DSKRVDPFVQKIERGDFFVPEFENLGMQPLFAKNISYKYNNNTANSRI 469
Query 589 ------GKQPAWIEYMTNVNKTYGNFALVENEGWMCLNRIFGD----INTYTTYIFPHLY 638
G QP + EY T ++ +G F E + + R G+ N T I P
Sbjct 470 KNLGAFGWQPRYSEYKTALDINHGQFVHQEPLSYWTVARARGESMSNFNISTFKINPKWL 529
Query 639 NNIFA----DTDVTSQNF 652
+++FA T++T Q F
Sbjct 530 DDVFAVNYNGTELTDQVF 547
>gi|609718276|emb|CDN73650.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=537
Score = 65.1 bits (157), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 72/254 (28%), Positives = 111/254 (44%), Gaps = 23/254 (9%)
Query 377 EDISGRHIPNCAYPMVGLALKTYQSDVNTNW--VNTEWLDGDSGINSITAIDTSGGSFTL 434
+D++G PN K +SDVN N V+ + L D N + + S T+
Sbjct 243 KDMAGNPAPN----------KDLRSDVNGNLQDVSGQPLSLDPSKNLKLNMASENVS-TV 291
Query 435 DTLNLAKKVYTMLNRIAISDGSYNAWIQTVY---TSGGLNHIETPIYLGGSSLEIEFQEV 491
+ L A K+ L + A + Y I + + TS G ++ P +LGG+ I EV
Sbjct 292 NDLRRAFKLQEWLEKNARAGSRYAESILSFFGVKTSDG--RLQRPEFLGGNKSPIMISEV 349
Query 492 VNNSGTEDQ-PLGTLAGRGVATNHKGGNIVFKADEPGYLFCITSITPRVDYFQGNEWDMY 550
+ S T+ P G +AG G+ GG F +E GY+ + S+ P+ Y QG
Sbjct 350 LQQSATDSTTPQGNMAGHGIGIGKDGGFSRF-FEEHGYVIGLMSVIPKTSYSQGIPRHFS 408
Query 551 LESLDDLHKPQLDGIGFQDRLYKHINA-NTDSTEFNKTVGKQPAWIEYMTNVNKTYGNFA 609
D PQ + IG Q K I A N D+ + G P + EY + + +G+F
Sbjct 409 KSDKFDYFWPQFEHIGEQPVYNKEIFAKNIDAFDSEAVFGYLPRYSEYKFSPSTVHGDFK 468
Query 610 LVENEGWMCLNRIF 623
++ + L RIF
Sbjct 469 --DDLYFWHLGRIF 480
Lambda K H a alpha
0.317 0.134 0.400 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 5172646292496