bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
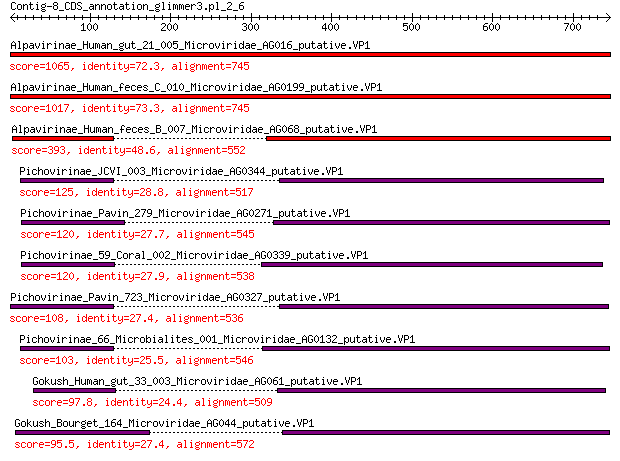
Query= Contig-8_CDS_annotation_glimmer3.pl_2_6
Length=745
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_gut_21_005_Microviridae_AG016_putative.VP1 1065 0.0
Alpavirinae_Human_feces_C_010_Microviridae_AG0199_putative.VP1 1017 0.0
Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1 393 3e-126
Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1 125 3e-32
Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1 120 1e-30
Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1 120 1e-30
Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1 108 2e-26
Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1 103 5e-25
Gokush_Human_gut_33_003_Microviridae_AG061_putative.VP1 97.8 4e-23
Gokush_Bourget_164_Microviridae_AG044_putative.VP1 95.5 2e-22
> Alpavirinae_Human_gut_21_005_Microviridae_AG016_putative.VP1
Length=742
Score = 1065 bits (2753), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 539/751 (72%), Positives = 608/751 (81%), Gaps = 15/751 (2%)
Query 1 MAQNVFDATFDANNRIDVNSFDWSHVNNLTTNFGRITPVFCELVPAKGSLRINPEFGLEL 60
MAQN+FDATFDANNRIDVNSFDWSHVNNLTT+FGRITPVFCELVPAKGSLRINPEFGLEL
Sbjct 1 MAQNIFDATFDANNRIDVNSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLEL 60
Query 61 MPMVFPVQTRMFARLNFFKVTLRSMWEDYPDFISNFRDDLEEPYILPDKHGFERMLKTNT 120
MPMVFPVQTRMFARLNFFKVTLRSMWEDY DFISNFRDDLEEPYI + F +M T +
Sbjct 61 MPMVFPVQTRMFARLNFFKVTLRSMWEDYSDFISNFRDDLEEPYINCSEISFPKMFTTGS 120
Query 121 LGDYLGIPTQRTTFSNYSSSAINHCTPSGSSTSEGFYFVSPSLSWDDIYSQ--WTGTVGS 178
LGDYLG+PT++ S +SSS + S S+S+ VSPS SWD I S + + +
Sbjct 121 LGDYLGVPTRK---SYHSSSTYISRSMSACSSSKKIALVSPSYSWDTIISNLSFNSAIST 177
Query 179 VKTIGCVVSPSNEESTVFLLSNSLAIGKYTSASLSFKMTTTASFKKMPCRIIIYHTALKT 238
+ + C + + + S+ + + +Y +AS+ F + + +++ T T
Sbjct 178 IPGVSCADAYDKSSTVILKTSSPVKVQRYKNASVEFYLLNDVKRDYVSHGLLVGITPGNT 237
Query 239 KFYVDSEFALSDDNK--TARLSIPLGNVAEKLG--AIGSTVSEFCNMCilisnsslnsaa 294
+ EF + K R+ + LGN+A A + +F + SL
Sbjct 238 SIHT-WEFNAPVNTKGNLTRMVVTLGNLANVFSDQAADADGIDFYVLLPSADMPSLTD-- 294
Query 295 naVTAQSVNVYADPELSEVDYDTYPFRTQKNDTAEHPKLLAYRFRAYESVYNAYYRDIRN 354
+ ++ + PE SE+ +D +PF++Q+ T +P+LLAYRFRAYE+VYNAYYRDIRN
Sbjct 295 --IALSTLTFFESPEESEITFDDFPFKSQEAGTKSYPRLLAYRFRAYEAVYNAYYRDIRN 352
Query 355 NPFVVNGRPVYNKWLPTMKGGADSTLYQLHQCNWERDFLTTAVPNPQQGMNTPLVGLTIG 414
NPFVVNGRPVYNKWLPTMKGGAD T+YQLHQCNWERDFLTTAVPNPQQG N PLVGLT+G
Sbjct 353 NPFVVNGRPVYNKWLPTMKGGADETMYQLHQCNWERDFLTTAVPNPQQGANAPLVGLTVG 412
Query 415 DVVTRSEDGTLSVQKQTVLVDEDGSKYGVSYRVSEDGERLVGVDYDPVSEKTPVTAINSY 474
DVVTRSEDGTLSVQKQTVLVDEDGSKYG+SY+VSEDGERLVGVDYDPVSEKTPVTAINSY
Sbjct 413 DVVTRSEDGTLSVQKQTVLVDEDGSKYGISYKVSEDGERLVGVDYDPVSEKTPVTAINSY 472
Query 475 aelaalaaeQSAGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPE 534
AELAALA EQ +GFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPE
Sbjct 473 AELAALATEQGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPE 532
Query 535 FIGGISRELSMRTVEQTVDQQGSSSQGQYAEALGSKTGIAGVYGNTSNNIEVFCDEESYI 594
FIGGISRELSMRTVEQTVDQQ +SSQGQYAEALGSKTGIAGVYG+TSNNIEVFCDEESYI
Sbjct 533 FIGGISRELSMRTVEQTVDQQSASSQGQYAEALGSKTGIAGVYGSTSNNIEVFCDEESYI 592
Query 595 IGLLTVTPVPIYTQLLPKDFTYNGLLDHYQPEFDRIGFQPITYKEICPMNADNSTSSGFI 654
IGLLTVTPVPIY+QLLPKDFTYNGLLDHYQPEFDRIGFQPITYKE+CP+N D S + +
Sbjct 593 IGLLTVTPVPIYSQLLPKDFTYNGLLDHYQPEFDRIGFQPITYKEVCPLNFDISNMND-M 651
Query 655 ERTFGYQRPWYEYVAKYDNAHGLFRTDMKNFVMHRSFTGLPQLGQQFLLVDPNAVNQVFS 714
+TFGYQRPWYEYVAKYDNAHGLFRTD+K FVMHR+F+GLPQLGQQFLLVDP+ VNQVFS
Sbjct 652 NKTFGYQRPWYEYVAKYDNAHGLFRTDLKKFVMHRTFSGLPQLGQQFLLVDPDTVNQVFS 711
Query 715 VTEYTDKIFGYVKFNATARLPISRVAIPRLD 745
VTEYTDKIFGYVKFNATARLPISRVAIPRLD
Sbjct 712 VTEYTDKIFGYVKFNATARLPISRVAIPRLD 742
> Alpavirinae_Human_feces_C_010_Microviridae_AG0199_putative.VP1
Length=731
Score = 1017 bits (2629), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 546/758 (72%), Positives = 593/758 (78%), Gaps = 40/758 (5%)
Query 1 MAQNVFDATFDANNRIDVNSFDWSHVNNLTTNFGRITPVFCELVPAKGSLRINPEFGLEL 60
MAQN+FDATFDANNRIDVNSFDWSHVNNLTT+FGRITPVFCELVPAKGSLRINPEFGLEL
Sbjct 1 MAQNIFDATFDANNRIDVNSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLEL 60
Query 61 MPMVFPVQTRMFARLNFFKVTLRSMWEDYPDFISNFRDDLEEPYILPDKHGFERMLKTNT 120
MPMVFPVQTRMFARLNFFKVTLRSMWEDY DFISNFRDDLEEPYILPD FE+M KT +
Sbjct 61 MPMVFPVQTRMFARLNFFKVTLRSMWEDYSDFISNFRDDLEEPYILPDSFRFEKMCKTGS 120
Query 121 LGDYLGIPTQRTTFSNYSSSAINHC-----TPSGSSTSEGFYFVSPSLSWDDIYSQWTGT 175
LGDYLG+PT T S + N C T S S + V S D T T
Sbjct 121 LGDYLGLPTFGTKSSRVYNFNDNPCAARSRTVSKSDIGKVMASVRSSSGIPDSVCASTLT 180
Query 176 VGSVKTIGCVVSPSNEESTVFLLSNSLAIGKYTSASLSFKMTTTASFKKMPCRIIIYHTA 235
G KT VS ++ S+ + S+ + IG + S +A+F
Sbjct 181 PGVSKT----VSFTSVNSSNVVPSDIIQIGIDITISGG----DSAAFDG----------- 221
Query 236 LKTKFYVDSEFALSDDNKTARLSIPLGNVAEK---LGAIGST-VSEFCNMCilisnssln 291
T ++V LS DNK IP+ AEK + I S S+F + +
Sbjct 222 -TTGYFV----LLSKDNKYL-FDIPITLKAEKGVVMAHISSVEASKFGSSYGTPLVIATP 275
Query 292 saanaVTAQSVN---VYADPEL-SEVDYDTYPFRTQKNDTAEHPKLLAYRFRAYESVYNA 347
+ ++ ++ VY L S+V Y TYPF T+ + +LLAYRFRAYESVYNA
Sbjct 276 DTTDVISFDNIKFSVVYTTYSLQSDVSYLTYPFATESR--PDISRLLAYRFRAYESVYNA 333
Query 348 YYRDIRNNPFVVNGRPVYNKWLPTMKGGADSTLYQLHQCNWERDFLTTAVPNPQQGMNTP 407
YYRDIRNNPFVVNGRPVYNKWLPTMKGGAD+TLY+L QCNWERDFLTTAVPNPQQG N P
Sbjct 334 YYRDIRNNPFVVNGRPVYNKWLPTMKGGADTTLYELRQCNWERDFLTTAVPNPQQGANAP 393
Query 408 LVGLTIGDVVTRSEDGTLSVQKQTVLVDEDGSKYGVSYRVSEDGERLVGVDYDPVSEKTP 467
LVGL +GDVVTRSEDGT SVQKQTVLVDEDGSKYGVSY+VSEDGERLVGVDYDPVSEKTP
Sbjct 394 LVGLVMGDVVTRSEDGTYSVQKQTVLVDEDGSKYGVSYKVSEDGERLVGVDYDPVSEKTP 453
Query 468 VTAINSYaelaalaaeQSAGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRF 527
VTAI SYAELAALA EQ +GFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRF
Sbjct 454 VTAIGSYAELAALATEQGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRF 513
Query 528 DELLMPEFIGGISRELSMRTVEQTVDQQGSSSQGQYAEALGSKTGIAGVYGNTSNNIEVF 587
DELLMPEFIGGISRELSMRTVEQTVDQQ SS+GQYAEALGSKTGIAGVYG+TSNNIEVF
Sbjct 514 DELLMPEFIGGISRELSMRTVEQTVDQQSGSSRGQYAEALGSKTGIAGVYGSTSNNIEVF 573
Query 588 CDEESYIIGLLTVTPVPIYTQLLPKDFTYNGLLDHYQPEFDRIGFQPITYKEICPMNADN 647
CDEESYIIGLLTVTPVPIYTQLLPKDFTYNGLLDHYQPEFDRIGFQPITYKEICPMN
Sbjct 574 CDEESYIIGLLTVTPVPIYTQLLPKDFTYNGLLDHYQPEFDRIGFQPITYKEICPMNIVA 633
Query 648 STSSGFIERTFGYQRPWYEYVAKYDNAHGLFRTDMKNFVMHRSFTGLPQLGQQFLLVDPN 707
S+ + +TFGYQRPWYEYVAKYD+AHGLFRT+MKNF+M R F+GLPQLGQQFLLVDP+
Sbjct 634 GDSANQLNKTFGYQRPWYEYVAKYDSAHGLFRTNMKNFIMSRVFSGLPQLGQQFLLVDPD 693
Query 708 AVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD 745
VNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD
Sbjct 694 TVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRLD 731
> Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1
Length=780
Score = 393 bits (1010), Expect = 3e-126, Method: Compositional matrix adjust.
Identities = 198/428 (46%), Positives = 285/428 (67%), Gaps = 16/428 (4%)
Query 319 PFRTQKNDTAEHPKLLAYRFRAYESVYNAYYRDIRNNPFVVNGRPVYNKWLPTMKGGADS 378
P+ + N T + K+ AY FRAY+++YNAY R+ RNNPF++NG+ YN+W+ T +GG DS
Sbjct 368 PYYSVYNTTEKAIKVSAYPFRAYQAIYNAYIRNTRNNPFILNGKKTYNRWITTDEGGEDS 427
Query 379 -TLYQLHQCNWERDFLTTAVPNPQQGMNTPLVGLTIGDVVTRSEDGTLSVQKQTVLVDED 437
T L NW+ D TTA+ PQQG+ PLVGLT + V+ ++ G T +VDED
Sbjct 428 LTPLDLLYANWQSDAYTTALTAPQQGV-APLVGLTTYESVSLNDAGHTVTTVNTAIVDED 486
Query 438 GSKYGVSYRVSEDGERLVGVDYDPVSEKTPVTAINSYaelaalaaeQSAGFTIETLRYVN 497
G+ Y V + +GE L GV+Y P+ AIN + +L + ++G +I R VN
Sbjct 487 GNAYKVDFE--SNGEALKGVNYTPLKAGE---AIN----MQSLVSPVTSGISINDFRNVN 537
Query 498 AYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQGS 557
AYQ++LELN +GFSYK+I++GR+D+++R+D L MPE++GGI+R++ + + QTV+ +
Sbjct 538 AYQRYLELNQFRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIVVNPITQTVE---T 594
Query 558 SSQGQYAEALGSKTGIAGVYGNTSNNIEVFCDEESYIIGLLTVTPVPIYTQLLPKDFTYN 617
+ G Y +LGS+ G+A +GN+ +I VFCDEES ++G++ V P+P+Y +LPK TY
Sbjct 595 TDTGSYVGSLGSQAGLATCFGNSDGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYR 654
Query 618 GLLDHYQPEFDRIGFQPITYKEICPMNADNSTSSGFIERTFGYQRPWYEYVAKYDNAHGL 677
LD + PEFD IG+QPI KE+ + A S + FGYQRPWYEYV K D AHGL
Sbjct 655 ERLDSFNPEFDHIGYQPIYLKELAAIQAFESGKD--LNTVFGYQRPWYEYVQKVDRAHGL 712
Query 678 FRTDMKNFVMHRSFTGLPQLGQQFLLVDPNAVNQVFSVTEYTDKIFGYVKFNATARLPIS 737
F + ++NF+M RSF P+LG+ F ++ P +VN VFSVTE +DKI G + F+ TA+LPIS
Sbjct 713 FLSSLRNFIMFRSFENAPELGKSFTVMQPGSVNNVFSVTEVSDKILGQIHFDCTAQLPIS 772
Query 738 RVAIPRLD 745
RV +PRL+
Sbjct 773 RVVVPRLE 780
Score = 137 bits (345), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 70/130 (54%), Positives = 87/130 (67%), Gaps = 7/130 (5%)
Query 4 NVFDATFDANNRIDVNSFDWSHVNNLTTNFGRITPVFCELVPAKGSLRINPEFGLELMPM 63
+VF+ D N + NSFDWSH NN TT+ GRITPVF ELVP S+RI PEFGL MPM
Sbjct 3 SVFNKIGDVKNDVKRNSFDWSHDNNFTTDLGRITPVFTELVPPNSSVRIKPEFGLRFMPM 62
Query 64 VFPVQTRMFARLNFFKVTLRSMWEDYPDFISNFRDDLEE---PYILPDKHGFER--MLKT 118
+FP+QT+M A L+FFKV LR++W+DY DFIS D+ EE P+I F L
Sbjct 63 MFPIQTKMKAYLSFFKVPLRTLWKDYMDFIS--ADNTEEYQPPFIKFSGKSFSEGGALSE 120
Query 119 NTLGDYLGIP 128
+ LGDY+G+P
Sbjct 121 SGLGDYIGMP 130
> Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1
Length=505
Score = 125 bits (315), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 118/413 (29%), Positives = 182/413 (44%), Gaps = 60/413 (15%)
Query 335 AYRFRAYESVYNAYYRD---IRNNPFVVNGRPVYNKWLPTMKGGADSTLYQLHQCNWERD 391
A F AY+ VYN +YRD + P+ + P L M+ A +E D
Sbjct 136 ALPFAAYQCVYNEWYRDQNLVAEVPYQLINGPNPRTNLAVMRLRA-----------FEHD 184
Query 392 FLTTAVPNPQQGMNTPLVGLTIGDVVTRSE---DGTLSVQKQTVLVDEDGSKYGVSYRVS 448
+ T+A+P Q+G V + +GD+ S+ DG SV K E+ + VS V+
Sbjct 185 YFTSALPFAQKGN---AVDIPLGDITLNSDWYLDG--SVPKL-----ENSTGGYVSGNVT 234
Query 449 ED---GERLVGVDYDPVSEKTPVTAINSYaelaalaaeQSAGFTIETLRYVNAYQKFLEL 505
D GE VG D DP + + + TI LR Q++LEL
Sbjct 235 SDNATGEINVGSDNDPFAYDP-------------DGSLEVTSTTITDLRRAFKLQEWLEL 281
Query 506 NMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQGSSSQGQYAE 565
R G Y + ++ +D+ L PE+I G + + + + T + QG
Sbjct 282 LARGGSRYIEQIKTFFDVKSSDQRLQRPEYITGTKQPVIISEILNTTGETAGLPQGNM-- 339
Query 566 ALGSKTGIAGVYGNTSNNIEVFCDEESYIIGLLTVTPVPIYTQLLPKDFTYNGLLDHYQP 625
S G+A GN + +C+E +IIG+L+VTP Y Q +PK + D Y P
Sbjct 340 ---SGHGVAVGTGNVG---KYYCEEHGFIIGILSVTPNTAYQQGIPKHYLRTDKYDFYWP 393
Query 626 EFDRIGFQPITYKEICPMNADNSTSSGFIERTFGYQRPWYEYVAKYDNAHGLFRTDMKNF 685
+F IG QPI EI + + TFGY + EY G FR+ + +
Sbjct 394 QFAHIGEQPIVNNEIYAYDPTG-------DETFGYTPRYAEYKYMPSRVAGEFRSSLDEW 446
Query 686 VMHRSFTGLPQLGQQFLLVDPNAVNQVFSVTEYTDKIFGYV--KFNATARLPI 736
R F+ LP L Q F+ VD + +++F+VT+ D ++ +V K A ++P+
Sbjct 447 HAARIFSSLPTLSQDFIEVDADDFDRIFAVTDGDDNLYMHVLNKVKARRQMPV 499
Score = 43.5 bits (101), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 31/129 (24%), Positives = 61/129 (47%), Gaps = 26/129 (20%)
Query 14 NRIDV-----NSFDWSHVNNLTTNFGRITPVF-CELVPAKGSLRINPEFGLELMPMVFPV 67
NR+ V N+FD +H ++ G + P+ E +P ++ + + ++ P++ PV
Sbjct 8 NRVSVKKERYNTFDLTHDVKMSGRMGELLPIMNLECIPGD-NITLGADSMVKFAPLLAPV 66
Query 68 QTRMFARLNFFKVTLRSMWEDYPDFISNFRDDLEEPYILPDKHGFERMLKTNT------- 120
R +++F V R +W+ + DF++ D++ P P +++ N+
Sbjct 67 MHRFNVTMHYFFVPNRIIWDGWEDFMT----DVDTPPAFP-------IIEVNSGWTAAQQ 115
Query 121 -LGDYLGIP 128
DYLGIP
Sbjct 116 RFADYLGIP 124
> Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1
Length=503
Score = 120 bits (302), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 119/428 (28%), Positives = 180/428 (42%), Gaps = 62/428 (14%)
Query 328 AEHPKLLAYRFRAYESVYNAYYRDIRNNPFVVNGRPVYNKWLPTMKGGADSTLYQLHQCN 387
A ++ A F AY+++YN YYRD N VN YN L G T L +
Sbjct 127 ATQVQINALPFAAYQAIYNEYYRD-ENLVTAVN----YN--LADGNQGI-GTFGTLRKRA 178
Query 388 WERDFLTTAVPNPQQGMNTPLVGLTIGDVVTRSEDGTLSVQK-----QTVLVDEDGSKYG 442
+E D+ T +P Q+G V L +GDV ++ ++ K T + + D + G
Sbjct 179 YEHDYFTACLPFAQKGA---AVDLPLGDVTLKANWNPSAIPKFVNAAGTSINNLDDVQGG 235
Query 443 VSYRVSEDGERLVGVDYDPVSEKTPVTAINSYaelaalaaeQSAGFTIETLRYVNAYQKF 502
R+ G + + Y+P + + TI LR Q+F
Sbjct 236 TPNRIRTSGGQ-SPIAYNP------------------NGSLEVGSTTINDLRRAMRLQEF 276
Query 503 LELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQGSSSQGQ 562
LE N R G Y + ++ + + L PE+I G + ++ + TV
Sbjct 277 LEKNARGGTRYIENIKMHFGVQSSDSRLQRPEYITGTKAPVVVQEITSTVKG-------- 328
Query 563 YAEALGSKTGIA-----GVYGNTSNNIEVFCDEESYIIGLLTVTPVPIYTQLLPKDFTYN 617
E G+ TG A G YGN S +E YIIG++++ P P Y Q +PK +
Sbjct 329 IEEPQGNPTGKATSLQSGHYGNYS------VEEHGYIIGIMSIMPKPAYQQGIPKSYLKT 382
Query 618 GLLDHYQPEFDRIGFQPITYKEICPMNADNSTSSGFIERTFGYQRPWYEYVAKYDNAHGL 677
+LD Y P F IG Q + +EI + G+I R + EY + GL
Sbjct 383 DVLDFYWPSFANIGEQEVLQEEIYGYTPTPNAVFGYIPR-------YAEYKYLQNRVCGL 435
Query 678 FRTDMKNFVMHRSFTGLPQLGQQFLLVDPNAVNQVFSVTE-YTDKIFGYVKFNATARLPI 736
FRT + + + R F +P L Q F+ V P ++F+ E Y D I+ +V TAR P+
Sbjct 436 FRTTLDFWHLGRIFANIPTLSQSFIEVSPANQTRIFANQEDYDDNIYIHVLNKMTARRPM 495
Query 737 SRVAIPRL 744
P L
Sbjct 496 PVFGTPML 503
Score = 37.7 bits (86), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 32/131 (24%), Positives = 54/131 (41%), Gaps = 10/131 (8%)
Query 15 RIDVNSFDWSHVNNLTTNFGRITPV-FCELVPAKGSLRINPEFGLELMPMVFPVQTRMFA 73
R N+FD +H +T G + P E VP I+ + L P+ P+ +
Sbjct 12 RPPSNTFDLTHDVKMTGGMGNLMPCCIAECVPGD-KWNISGDTYLRFAPITAPIMHSIDV 70
Query 74 RLNFFKVTLRSMWEDYPDFISNFRDDLEEPYILPDKHGFERMLKTNT--LGDYLGIPTQR 131
+++F V R +W ++ FI N EP ++ + LGDYL +P
Sbjct 71 AIHYFFVPNRILWSNWEKFIVN------EPTGGIPTFSWDANIPATAKKLGDYLSVPPPP 124
Query 132 TTFSNYSSSAI 142
T + +A+
Sbjct 125 TGATQVQINAL 135
> Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1
Length=522
Score = 120 bits (302), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 115/444 (26%), Positives = 185/444 (42%), Gaps = 68/444 (15%)
Query 313 VDYDTYPFRTQKNDTAEHPKLLAYRFRAYESVYNAYYRDIRNNPFVVNGRPVYNKWLPTM 372
DY P TQ + + P F AY+ +Y+ YYRD + +K T+
Sbjct 119 ADYLGLPTGTQIDGVSALP------FAAYQKIYDDYYRD----------ENLIDKVDITL 162
Query 373 ----KGGADST-LYQLHQCNWERDFLTTAVPNPQQGMNTPLVGLTIGDVVTRSEDGTLSV 427
+ GAD+ L + + W+ D+ T+A+P Q+G + T+ +
Sbjct 163 SDGTQSGADAIELGTMRKRAWQHDYFTSALPWTQRG-----------------PEATIPL 205
Query 428 QKQTVLVDEDGSKYGVSYRVSEDGERLVGVDYDPVSE-------------KTPVTAINSY 474
+ + + R + G + G +D S TPV+
Sbjct 206 GTSAPITWSNNTATATIVRNNTSGNPIAGYTFDGASALYTGGSGQLIADLPTPVSLDFDN 265
Query 475 aelaalaaeQSAGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPE 534
++ + +I LR Q++LE N R G Y +I+ + + L PE
Sbjct 266 SDHLFADLAGATASSINDLRRAFRLQEWLERNARGGARYIEIIMAHFGVRSSDARLQRPE 325
Query 535 FIGGISRELSMRTVEQT-VDQQGSSSQGQYAEALGSKTGIAGVYGNTSNNIEVFCDEESY 593
F+GG + +++ V QT + + QG A GV +SN + C+E Y
Sbjct 326 FLGGSATPITISEVLQTSANNTEPTPQGNMAGH--------GVSVGSSNYVSYKCEEHGY 377
Query 594 IIGLLTVTPVPIYTQLLPKDFTYNGLLDHYQPEFDRIGFQPITYKEICPMNADNSTSSGF 653
IIG+++V P Y Q +PK F D+Y P F IG QPI +E+ N N+T +
Sbjct 378 IIGIMSVMPKTAYQQGIPKHFKKLDKFDYYWPSFANIGEQPILNEELYHQN--NATDT-- 433
Query 654 IERTFGYQRPWYEYVAKYDNAHGLFRTDMKNFVMHRSFTGLPQLGQQFLLVDPNAVNQVF 713
TFGY + EY HG FR + + M R F P L Q F+ + + V +VF
Sbjct 434 --ETFGYTPRYAEYKYIPSTVHGEFRDSLDFWHMGRIFGSKPTLNQDFIECNADDVERVF 491
Query 714 SVTEYTDKIFGYV--KFNATARLP 735
+VT + ++ Y+ + AT +P
Sbjct 492 AVTAGQEHLYVYLHNEVKATRLMP 515
Score = 51.2 bits (121), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 35/121 (29%), Positives = 51/121 (42%), Gaps = 11/121 (9%)
Query 15 RIDVNSFDWSHVNNLTTNFGRITPV-FCELVPAKGSLRINPEFGLELMPMVFPVQTRMFA 73
R N+FD SH + G + P+ E VP I P++ P+ +
Sbjct 11 RPQTNTFDLSHDRKFSGKIGELMPITVMEAVPGD-KFNIKATNLTRFAPLITPIMHQASV 69
Query 74 RLNFFKVTLRSMWEDYPDFISNFRDDLEEPY-----ILPDKHGFERMLKTNTLGDYLGIP 128
+FF V R +W ++ DFIS D L +P I +G+ L DYLG+P
Sbjct 70 YCHFFFVPNRILWSNWEDFISGGEDGLADPTFPTINITTPTNGY----AVGALADYLGLP 125
Query 129 T 129
T
Sbjct 126 T 126
> Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1
Length=508
Score = 108 bits (269), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 107/413 (26%), Positives = 165/413 (40%), Gaps = 49/413 (12%)
Query 335 AYRFRAYESVYNAYYRDIRNNPFVVNGRPVYNKWLP---TMKGGADSTLYQLHQCNWERD 391
A AY+ +YN YYRD PV K G L L Q WE D
Sbjct 137 ALPMAAYQCIYNEYYRDQNLQ------TPVDYKLTDGNNNTDAGDRERLTTLRQRAWEHD 190
Query 392 FLTTAVPNPQQGMNTPLVGLTIGDVVTRSEDGTLSVQKQTVLVDEDGSKYGVSYRVSEDG 451
+ T ++P Q+G V + IG + D ++V ++ +Y S DG
Sbjct 191 YFTASLPFAQKGA---AVDIPIGTI-----DKDVAVNFNSLEFGATTVRYN-----SADG 237
Query 452 ERLVGVDYDPVSEKTPVTAINSYaelaalaaeQSAGFTIETLRYVNAYQKFLELNMRKGF 511
+G ++ TP + TI LR Q++LE N R G
Sbjct 238 LIDIGGSNAVNADGTPTMIAKTSTIDIEPT-------TINDLRRAFKLQEWLEKNARGGT 290
Query 512 SYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQGSSSQGQYA-EALGSK 570
Y + + + + L PE+I G+ + + V T Q G QG A +
Sbjct 291 RYIENILTHFGVRSSDKRLQRPEYITGVKSPVVVSEVLNTTGQDGGLPQGNMAGHGISVT 350
Query 571 TGIAGVYGNTSNNIEVFCDEESYIIGLLTVTPVPIYTQLLPKDFTYNGLLDHYQPEFDRI 630
+G +G Y +C+E YIIG+++V P Y Q +P+ F LD++ P F I
Sbjct 351 SGKSGSY---------YCEEHGYIIGIMSVMPKTAYQQGIPRTFLKTDSLDYFWPTFANI 401
Query 631 GFQPITYKEICPMNADNSTSSGFIERTFGYQRPWYEYVAKYDNAHGLFRTDMKNFVMHRS 690
G Q + +E+ A+ + TFGY + EY G FRT + + + R
Sbjct 402 GEQEVAKQELYAYTANAND-------TFGYVPRYAEYKYMPSRVAGEFRTSLNYWHLGRI 454
Query 691 FTGLPQLGQQFLLVDPNAVNQVFSVTE-YTDKIFGYVKFNATARLPISRVAIP 742
F P L F+ DP ++F+V + TD ++ +V A P+ + P
Sbjct 455 FATEPSLNSDFIECDP--TKRIFAVEDPETDVLYCHVLNKIKAVRPMPKYGTP 505
Score = 63.2 bits (152), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 40/129 (31%), Positives = 64/129 (50%), Gaps = 6/129 (5%)
Query 1 MAQNVFDATFDANNRIDVNSFDWSHVNNLTTNFGRITPVF-CELVPAKGSLRINPEFGLE 59
M QN+F++ N+ N FD +H L+TN G++TP+ E VP ++ E +
Sbjct 1 MGQNLFNSI--QLNKPKKNVFDLTHDVKLSTNMGQLTPILTLECVPGD-KFDLSCESLIR 57
Query 60 LMPMVFPVQTRMFARLNFFKVTLRSMWEDYPDFISNFRDDLEEPYILPDKHGFERMLKTN 119
PM+ PV RM +++F V R +W ++ FI+ + PY+ + M K
Sbjct 58 FAPMIAPVMHRMDVTMHYFFVPNRILWSNWEKFITEHNSEHVAPYMAYTNGDYTAMQK-- 115
Query 120 TLGDYLGIP 128
DY+GIP
Sbjct 116 KFMDYIGIP 124
> Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1
Length=518
Score = 103 bits (258), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 105/441 (24%), Positives = 185/441 (42%), Gaps = 51/441 (12%)
Query 314 DYDTYPFRTQKNDTAEHPKLLAYRFRAYESVYNAYYRD--IRNNPFVVNGRPVYNKWLPT 371
D+ YP + +D P LA AY+ +++ YYRD +++ F P
Sbjct 119 DHLGYPLSSVSSDVKASPMPLA----AYQKIWDEYYRDQNLQDERFT-----------PL 163
Query 372 MKGGADSTLYQLHQC---NWERDFLTTAVPNPQQGMNTPLVGLTIGDVVTRSEDGTLSVQ 428
+ G +S L QC W D+ T+ +P Q+G + L + + + + D T +
Sbjct 164 VPGDNNSQLALNQQCYRRAWMHDYFTSCLPFAQKGDSVQLPISSAENQIVQL-DPTATAP 222
Query 429 KQTVLVDEDGSKYGVSYRVSEDG----ERLVGVDYDPVSEKTPVTAINSYaelaalaaeQ 484
VL D S + + + +V D +P+ + + + Q
Sbjct 223 GTFVLADGQASATPGNVHIINGPPPYVQTVVDDDDNPLQFEPRGSLV---------VDVQ 273
Query 485 SAGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELS 544
+ I T+R Q++LE N R G Y + + + + L PE++GG +
Sbjct 274 ADAVDINTVRRAFRLQEWLEKNARAGTRYIESILSHFGVRSSDARLQRPEYLGGTKGNMV 333
Query 545 MRTVEQTVDQQGSSSQGQYAEALGSKTGIAGVYGNTSNNIEVFCDEESYIIGLLTVTPVP 604
+ V T + ++ A+G+ G G+ + N I+ C+E +IIG++ V PV
Sbjct 334 ISEVLATAETTDANV------AVGTMAG-HGITAHGGNTIKYRCEEHGWIIGIINVQPVT 386
Query 605 IYTQLLPKDFTYNGLLDHYQPEFDRIGFQPITYKEICPMNADNSTSSGFIERTFGYQRPW 664
Y Q LP++F+ LD+ P F IG Q + KE+ +S + FGY +
Sbjct 387 AYQQGLPREFSRFDRLDYPWPVFANIGEQEVYNKELY-------ANSPAPDEIFGYIPRY 439
Query 665 YEYVAKYDNAHGLFRTDMKNFVMHRSFTGLPQLGQQFLLVDPNAVNQVFSVTE-YTDKIF 723
E K G R + + + R F P L ++F+ +P+ ++F+VT+ D I
Sbjct 440 AEMKFKNSRVAGEMRDTLDFWHLGRIFNNTPALNEEFIECNPDT--RIFAVTDPAEDHIV 497
Query 724 GYVKFNATARLPISRVAIPRL 744
++ T + R IP +
Sbjct 498 AHIYNEVTVNRKLPRYGIPTI 518
Score = 55.5 bits (132), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 34/116 (29%), Positives = 60/116 (52%), Gaps = 7/116 (6%)
Query 14 NRIDVNSFDWSHVNNLTTNFGRITPV-FCELVPAKGSLRINPEFGLELMPMVFPVQTRMF 72
+++D N+FD SH + G++ P E +P I P+ L +P++ PV R+
Sbjct 15 SKVDSNNFDLSHDVKSSFKMGQLVPTNVIECMPGD-LFTIRPQNMLRFLPLIAPVMHRVE 73
Query 73 ARLNFFKVTLRSMWEDYPDFISNFRDDLEEPYILPDKHGFERMLKTNTLGDYLGIP 128
++F V R +W D+ +I+ D+E PY + ++ ++ TLGD+LG P
Sbjct 74 VTTHYFFVPNRLLWADWEKWITG-ESDVEAPYF----NFIDQGIEVGTLGDHLGYP 124
> Gokush_Human_gut_33_003_Microviridae_AG061_putative.VP1
Length=545
Score = 97.8 bits (242), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 103/432 (24%), Positives = 187/432 (43%), Gaps = 46/432 (11%)
Query 332 KLLAYRFRAYESVYNAYYRDIR-NNPFVVNGRPVYNKWLPTMKGGADSTL---YQLHQC- 386
K+ A RAY V+N ++RD N V+ ++ + K + L Y+ +C
Sbjct 130 KVNALPVRAYVKVWNEFFRDENVQNAAVIKTDDADVQYEDSSKDNVEENLKVAYKGGRCL 189
Query 387 --NWERDFLTTAVPNPQQG--MNTPLVG---LTIGDVVTRSEDGTLSVQKQTVLVDEDGS 439
N D+ T+++P Q+G + P+ G + +G+ +D V+ +++D G+
Sbjct 190 PVNKFHDYFTSSLPFAQRGPEVTVPMTGNAPIRVGNAKGGYQDFRGPVE---MVIDAHGA 246
Query 440 K------YGVSYRVSEDGERLVGVDYDPVSEKTPVTAINSYaelaalaaeQSAGFTIETL 493
YG S + + ++ S K VT + TI L
Sbjct 247 DIPGSLIYGNSTGAPGEKKSMM------FSGKEKVTNEMGAGGWMYADISEVTAATINDL 300
Query 494 RYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVD 553
R A Q++ E R G Y++ +Q WD+ I + +PE++GG ++M + QT
Sbjct 301 RKAVAVQQYYEALARGGSRYREQVQALWDVVISDKTMQIPEYLGGGRYHVNMNQIVQTSG 360
Query 554 QQGSSSQGQYAEALGSKTGIAGVYGNTSNNIEVFCDEESYIIGLLTVTPVPIYTQLLPKD 613
QQ + +G ++ N S+ + F +E ++IG+ V Y Q L +
Sbjct 361 QQTNDDT-----PIGETGAMSVTPINESSFTKSF-EEHGFVIGVCCVRHNHSYQQGLERF 414
Query 614 FTYNGLLDHYQPEFDRIGFQPITYKEICPMNADNSTSSGFIERTFGYQRPWYEYVAKYDN 673
++ LD+Y P+F +G QPI KEI T + E TFGYQ W +Y K +
Sbjct 415 WSRTDRLDYYVPQFANLGEQPIKKKEIM------LTGTASDEETFGYQEAWADYRMKPNR 468
Query 674 AHGLFRTDMKN----FVMHRSFTGLPQLGQQFLLVDPNAVNQVFSVTEYTDKIFGYVKF- 728
GL R++ + + +++ +P L Q+++ + + + + + FG ++
Sbjct 469 VSGLMRSNAEGTLDFWHYADNYSTVPTLSQEWIAEGKEEIARTL-IVQNEPQFFGAIRIA 527
Query 729 -NATARLPISRV 739
T R+P+ V
Sbjct 528 NKTTRRMPLYSV 539
Score = 27.7 bits (60), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 21/108 (19%), Positives = 49/108 (45%), Gaps = 7/108 (6%)
Query 30 TTNFGRITPVFCELVPAKGSLRINPEFGLELMPMVFPVQTRMFARLNFFKVTLRSMWEDY 89
T + G++ P + + V + ++ + + +PV F +F R +W+++
Sbjct 17 TFDSGKLIPFYVDEVLPGDTFSVDTTAIIRMTTPKYPVMDDAFIDFYYFYCPNRILWDNF 76
Query 90 PDFISNFRDDLEEP---YILPDKH--GFER--MLKTNTLGDYLGIPTQ 130
F+ + P Y +P+ + G E+ + ++ DY+G+PT+
Sbjct 77 KYFMGEAEETPWMPAKTYKVPEINIKGSEKSPLPNERSILDYMGVPTK 124
> Gokush_Bourget_164_Microviridae_AG044_putative.VP1
Length=540
Score = 95.5 bits (236), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 111/418 (27%), Positives = 175/418 (42%), Gaps = 45/418 (11%)
Query 339 RAYESVYNAYYRD--IRNNPFVVNGRPVYNKWLPTMKGGADSTLYQLHQCNWERDFLTTA 396
RAY +YN ++RD ++N+ V G + +T Y + + D+ T+A
Sbjct 152 RAYNLIYNQWFRDENLQNSSVVDKG---------DGPDASPATNYTILRRGKRHDYFTSA 202
Query 397 VPNPQQGMNTPLVGLTIGDVVTRSEDGTLSVQKQTVLVDEDGSKYGVSYRVSEDGERLVG 456
+P PQ+G V + IG +G + T K G S +++++G+ G
Sbjct 203 LPWPQKGGTA--VTIPIGTSAPIKSNGGANFYAWTGSSGTSQLKVGTSGQLTQNGQANPG 260
Query 457 VDYDPVSEKTPVTAINSYaelaalaaeQSAGFTIETLRYVNAYQKFLELNMRKGFSYKQI 516
D + T + + TI LR QK LE + R G Y +I
Sbjct 261 SIVDQTWGPSGTT------QGLYADLSTATAATINQLRQSFQIQKLLERDARGGTRYTEI 314
Query 517 MQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQT--VDQQGSSS-QGQYAEALGSKTGI 573
++ + + L PE++GG S +S+ + QT Q G ++ QG A A+G T +
Sbjct 315 LRSHFGVTSPDARLQRPEYLGGGSTLVSISPIAQTSGTGQTGQATPQGNLA-AMG--TYL 371
Query 574 AGVYGNTSNNIEVFCDEESYIIGLLTVTPVPIYTQLLPKDFTYNGLLDHYQPEFDRIGFQ 633
A +G T + + E YIIG+++V Y Q L + ++ + D+Y P F +G Q
Sbjct 372 AQNHGFTHSFV-----EHGYIIGVVSVRADLTYQQGLRRHWSRSTRYDYYFPAFAMLGEQ 426
Query 634 PITYKEI--CPMNADNSTSSGFIERTFGYQRPWYEYVAKYDNAHGLFRTDMKNFV----M 687
I KEI N+DN+ FGYQ W EY GLFR+ +
Sbjct 427 AILNKEIYVTGSNSDNN--------VFGYQERWAEYRYNPSEITGLFRSTAAGTIDPWHY 478
Query 688 HRSFTGLPQLGQQFLLVDPN-AVNQVFSVTEYTDKIFGYVKFNATARLPISRVAIPRL 744
+ FT LP L F+ +P A N + FN A P+ ++P L
Sbjct 479 AQRFTSLPTLNSTFIQDNPPLARNLAVGSAANGQQFLLDAFFNINAARPLPMYSVPGL 536
Score = 45.1 bits (105), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 46/178 (26%), Positives = 73/178 (41%), Gaps = 32/178 (18%)
Query 7 DATFDANN-----RIDVNSFDWSHVNNLTTNF--GRITPVFCELVPAKGSLRINPEFGLE 59
+ + DA+N R D+ +S L T F G + P+ CE + + +N
Sbjct 4 NKSVDAHNFAMVPRADIPRSRFSMQKTLKTTFDSGLLVPIMCEEILPGDTFNVNVTMFGR 63
Query 60 LMPMVFPVQTRMFARLNFFKVTLRSMWEDYPDFIS---NFRDDLEEPYILPDKHGFERML 116
L +FPV + FF V R +W ++ F+ N D + Y +P +
Sbjct 64 LATPLFPVMDNLHLDSFFFFVPNRLVWTNWVKFMGEQDNPSDSIS--YTIPQQVSPAGGY 121
Query 117 KTNTLGDYLGIPT--QRTTFSNYSSSAINHCTPSGSSTSEGFYFVSPSLSWDDIYSQW 172
+L DYLG+PT Q T S S SA+ P+ +++ IY+QW
Sbjct 122 AIGSLQDYLGLPTVGQVTAGSTVSHSAL------------------PTRAYNLIYNQW 161
Lambda K H a alpha
0.319 0.135 0.405 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 69891720