bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
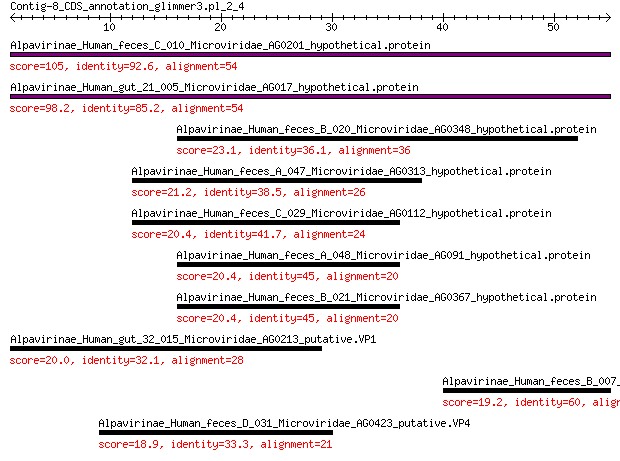
Query= Contig-8_CDS_annotation_glimmer3.pl_2_4
Length=54
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_C_010_Microviridae_AG0201_hypothetical.... 105 3e-33
Alpavirinae_Human_gut_21_005_Microviridae_AG017_hypothetical.pr... 98.2 2e-30
Alpavirinae_Human_feces_B_020_Microviridae_AG0348_hypothetical.... 23.1 0.10
Alpavirinae_Human_feces_A_047_Microviridae_AG0313_hypothetical.... 21.2 0.65
Alpavirinae_Human_feces_C_029_Microviridae_AG0112_hypothetical.... 20.4 1.0
Alpavirinae_Human_feces_A_048_Microviridae_AG091_hypothetical.p... 20.4 1.3
Alpavirinae_Human_feces_B_021_Microviridae_AG0367_hypothetical.... 20.4 1.4
Alpavirinae_Human_gut_32_015_Microviridae_AG0213_putative.VP1 20.0 1.6
Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1 19.2 4.2
Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4 18.9 5.6
> Alpavirinae_Human_feces_C_010_Microviridae_AG0201_hypothetical.protein
Length=54
Score = 105 bits (262), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 50/54 (93%), Positives = 53/54 (98%), Gaps = 0/54 (0%)
Query 1 MYKGVLKFIKRETLHEEFVISIGIFNRPSTAERFRKQLQDVNVGYDVLLILEKL 54
MYKGVLKFIKRETLHEEF+I+IGIFNRPSTAERFRKQLQD N+GYDVLLILEKL
Sbjct 1 MYKGVLKFIKRETLHEEFLINIGIFNRPSTAERFRKQLQDANIGYDVLLILEKL 54
> Alpavirinae_Human_gut_21_005_Microviridae_AG017_hypothetical.protein
Length=54
Score = 98.2 bits (243), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 46/54 (85%), Positives = 48/54 (89%), Gaps = 0/54 (0%)
Query 1 MYKGVLKFIKRETLHEEFVISIGIFNRPSTAERFRKQLQDVNVGYDVLLILEKL 54
MYK LKFIKRETL EEF+I IGIFNRPSTAERFRKQLQD N+GYDVLLI EKL
Sbjct 1 MYKASLKFIKRETLQEEFIIDIGIFNRPSTAERFRKQLQDANIGYDVLLIFEKL 54
> Alpavirinae_Human_feces_B_020_Microviridae_AG0348_hypothetical.protein
Length=81
Score = 23.1 bits (48), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 13/40 (33%), Positives = 23/40 (58%), Gaps = 6/40 (15%)
Query 16 EEFVISIGIFNRPSTAERFRK----QLQDVNVGYDVLLIL 51
EE++I+IG N +T E+F+ Q+Q +D++ L
Sbjct 17 EEYIITIG--NHLATEEKFKSAKAAQMQINKTNWDLVSAL 54
> Alpavirinae_Human_feces_A_047_Microviridae_AG0313_hypothetical.protein
Length=75
Score = 21.2 bits (43), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 10/26 (38%), Positives = 17/26 (65%), Gaps = 2/26 (8%)
Query 12 ETLHEEFVISIGIFNRPSTAERFRKQ 37
+T EF+I+IG N +T E+F+ +
Sbjct 14 DTDENEFIITIG--NHLATEEKFKSR 37
> Alpavirinae_Human_feces_C_029_Microviridae_AG0112_hypothetical.protein
Length=75
Score = 20.4 bits (41), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 10/24 (42%), Positives = 16/24 (67%), Gaps = 2/24 (8%)
Query 12 ETLHEEFVISIGIFNRPSTAERFR 35
+T EF+I+IG N +T E+F+
Sbjct 14 DTNENEFMITIG--NHLATEEKFK 35
> Alpavirinae_Human_feces_A_048_Microviridae_AG091_hypothetical.protein
Length=79
Score = 20.4 bits (41), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 9/20 (45%), Positives = 15/20 (75%), Gaps = 2/20 (10%)
Query 16 EEFVISIGIFNRPSTAERFR 35
+EF+I+IG N +T E+F+
Sbjct 17 DEFIITIG--NHLATEEKFK 34
> Alpavirinae_Human_feces_B_021_Microviridae_AG0367_hypothetical.protein
Length=79
Score = 20.4 bits (41), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 9/20 (45%), Positives = 15/20 (75%), Gaps = 2/20 (10%)
Query 16 EEFVISIGIFNRPSTAERFR 35
+EF+I+IG N +T E+F+
Sbjct 17 DEFIITIG--NHLATEEKFK 34
> Alpavirinae_Human_gut_32_015_Microviridae_AG0213_putative.VP1
Length=653
Score = 20.0 bits (40), Expect = 1.6, Method: Composition-based stats.
Identities = 9/28 (32%), Positives = 13/28 (46%), Gaps = 0/28 (0%)
Query 1 MYKGVLKFIKRETLHEEFVISIGIFNRP 28
MY + K T E F ++IG+ P
Sbjct 166 MYYDIFKNYFANTQEESFYVAIGVGEDP 193
> Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1
Length=780
Score = 19.2 bits (38), Expect = 4.2, Method: Composition-based stats.
Identities = 9/15 (60%), Positives = 10/15 (67%), Gaps = 0/15 (0%)
Query 40 DVNVGYDVLLILEKL 54
DVNV YD L + E L
Sbjct 562 DVNVRYDALNMPEYL 576
> Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4
Length=532
Score = 18.9 bits (37), Expect = 5.6, Method: Composition-based stats.
Identities = 7/21 (33%), Positives = 12/21 (57%), Gaps = 0/21 (0%)
Query 9 IKRETLHEEFVISIGIFNRPS 29
I++ H E ++GIF + S
Sbjct 505 IRKRIKHREINDAVGIFTKQS 525
Lambda K H a alpha
0.326 0.145 0.401 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3709748