bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
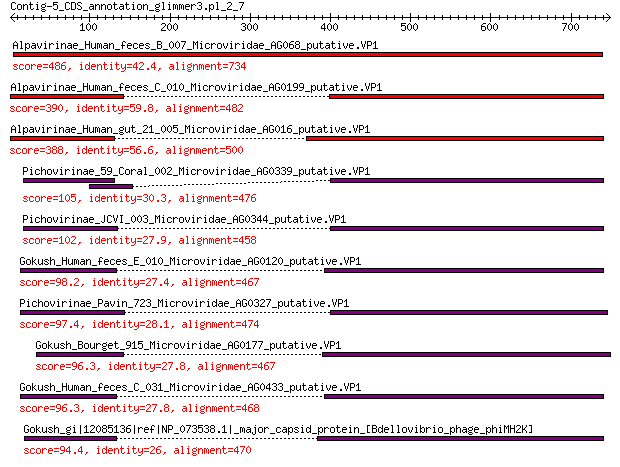
Query= Contig-5_CDS_annotation_glimmer3.pl_2_7
Length=748
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1 486 8e-162
Alpavirinae_Human_feces_C_010_Microviridae_AG0199_putative.VP1 390 1e-125
Alpavirinae_Human_gut_21_005_Microviridae_AG016_putative.VP1 388 9e-125
Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1 105 9e-26
Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1 102 1e-24
Gokush_Human_feces_E_010_Microviridae_AG0120_putative.VP1 98.2 4e-23
Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1 97.4 5e-23
Gokush_Bourget_915_Microviridae_AG0177_putative.VP1 96.3 1e-22
Gokush_Human_feces_C_031_Microviridae_AG0433_putative.VP1 96.3 1e-22
Gokush_gi|12085136|ref|NP_073538.1|_major_capsid_protein_[Bdell... 94.4 6e-22
> Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1
Length=780
Score = 486 bits (1251), Expect = 8e-162, Method: Compositional matrix adjust.
Identities = 311/769 (40%), Positives = 423/769 (55%), Gaps = 94/769 (12%)
Query 5 AFDATLDVNNEIKVNNFDWSHANNLTTQIGRVTPVFCELVPAHGSVRINPRFGLQFMPMV 64
F+ DV N++K N+FDWSH NN TT +GR+TPVF ELVP + SVRI P FGL+FMPM+
Sbjct 4 VFNKIGDVKNDVKRNSFDWSHDNNFTTDLGRITPVFTELVPPNSSVRIKPEFGLRFMPMM 63
Query 65 FPIQTRLRARMMFFKYPLRALWDGYRDFI-GNFREDLEEPYLDLN--TVTRLDAMAKTGS 121
FPIQT+++A + FFK PLR LW Y DFI + E+ + P++ + + + A++++G
Sbjct 64 FPIQTKMKAYLSFFKVPLRTLWKDYMDFISADNTEEYQPPFIKFSGKSFSEGGALSESG- 122
Query 122 LGDYLGLPTTLFGKFGTTLSVAT-------TGHIFGLKGNQFNGSDQ----MQYISG--V 168
LGDY+G+P + + V T +FG Q + + MQY++G
Sbjct 123 LGDYIGMPVSNIDTNWQVMDVGTLIDSTKGATAVFGSATKQGDLPELRFSVMQYVNGPFT 182
Query 169 PIDSVSNLRAFLSL-------KPL--------SIDTPSQRISWT--VVSPGSASSSETVN 211
P S S+ ++ + P+ +D + RI WT + G+++
Sbjct 183 PQISYSDNSCYMPIGNDYSFFSPIITFYFTKEMMDAGTFRIKWTGSLNIEGTSARQTAEQ 242
Query 212 IVPFVSASGIPGSDSITANSRLEFDIPWKRIGNPTSADLaafssgavagfaKKSTTYILM 271
V F S P I + + P IG P A F T Y +
Sbjct 243 FVNFTKCSKTP---VILTFKKKQTPAPVGDIGLPFDAS---QKVSFFEPFYVSDTIYRVK 296
Query 272 NFIQATFKVVN-GGRFVKLLFSFPESMYGSIPGDYEFFTFLNLYSAFGRRISGDSTAWGL 330
+ATF + N G + +F GSI Y+ F + ++ A G
Sbjct 297 FSFEATFNIANFPGMGINEDDAFSLGFTGSI---YDIFNLGGKLGSTDVNLTFSYNASGD 353
Query 331 QTTSFGRAFSGISDGEQTNSFTIRVSDFDLAGSGAQAHYYLTSDKPVDLTLGSSPYYNSG 390
+ F R SGIS +PYY S
Sbjct 354 SSLVFARPVSGIS----------------------------------------TPYY-SV 372
Query 391 SANKDKQIKISAYSFRAYEGIYNAYIRDNRNNPYYVNGQVQYNKWIPTYDGGADQ-NIYE 449
+K IK+SAY FRAY+ IYNAYIR+ RNNP+ +NG+ YN+WI T +GG D +
Sbjct 373 YNTTEKAIKVSAYPFRAYQAIYNAYIRNTRNNPFILNGKKTYNRWITTDEGGEDSLTPLD 432
Query 450 LRYANWEKDFLTTAVQSPQQGTAPLVGIttytetvettSDDGTPVTRELSRIALVDEDGK 509
L YANW+ D TTA+ +PQQG APLVG+TTY +D G VT A+VDEDG
Sbjct 433 LLYANWQSDAYTTALTAPQQGVAPLVGLTTYESVSL--NDAGHTVTT--VNTAIVDEDGN 488
Query 510 KYQVSFDSDSEGLKGVSYVELDNEVKLRQPRNLIDVVTSGISINDLRNVNAYQKFLELNM 569
Y+V F+S+ E LKGV+Y L + ++L+ VTSGISIND RNVNAYQ++LELN
Sbjct 489 AYKVDFESNGEALKGVNYTPLKAGEAINM-QSLVSPVTSGISINDFRNVNAYQRYLELNQ 547
Query 570 RKGYSYRDIIEGRFNVKVRYDELLMPEFFGGFSRDIEMHSISQTVDQDLDGSQTYAKALG 629
+G+SY++IIEGRF+V VRYD L MPE+ GG +RDI ++ I+QTV+ GS Y +LG
Sbjct 548 FRGFSYKEIIEGRFDVNVRYDALNMPEYLGGITRDIVVNPITQTVETTDTGS--YVGSLG 605
Query 630 SQSGIAGVRGDSGRALECFCDEESIVMGILIVTPLPVYTQLLPKHFTYRGLLDHYQPEFN 689
SQ+G+A G+S ++ FCDEESIVMGI+ V P+PVY +LPK TYR LD + PEF+
Sbjct 606 SQAGLATCFGNSDGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYRERLDSFNPEFD 665
Query 690 HIGFQPILYKEVCPLQAYSDGPDTLSDVFGYNRPWYEYVQKYDQAHGLF 738
HIG+QPI KE+ +QA+ G D L+ VFGY RPWYEYVQK D+AHGLF
Sbjct 666 HIGYQPIYLKELAAIQAFESGKD-LNTVFGYQRPWYEYVQKVDRAHGLF 713
> Alpavirinae_Human_feces_C_010_Microviridae_AG0199_putative.VP1
Length=731
Score = 390 bits (1003), Expect = 1e-125, Method: Compositional matrix adjust.
Identities = 195/352 (55%), Positives = 247/352 (70%), Gaps = 15/352 (4%)
Query 399 KISAYSFRAYEGIYNAYIRDNRNNPYYVNGQVQYNKWIPTYDGGADQNIYELRYANWEKD 458
++ AY FRAYE +YNAY RD RNNP+ VNG+ YNKW+PT GGAD +YELR NWE+D
Sbjct 318 RLLAYRFRAYESVYNAYYRDIRNNPFVVNGRPVYNKWLPTMKGGADTTLYELRQCNWERD 377
Query 459 FLTTAVQSPQQGT-APLVGIttytetvettSDDGTPVTRELSRIALVDEDGKKYQVSF-- 515
FLTTAV +PQQG APLVG+ S+DGT ++ + LVDEDG KY VS+
Sbjct 378 FLTTAVPNPQQGANAPLVGLVMGDVVTR--SEDGTYSVQK--QTVLVDEDGSKYGVSYKV 433
Query 516 DSDSEGLKGVSYVELDNEVKLRQPRNLIDVVT------SGISINDLRNVNAYQKFLELNM 569
D E L GV Y + + + + ++ SG +I LR VNAYQKFLELNM
Sbjct 434 SEDGERLVGVDYDPVSEKTPVTAIGSYAELAALATEQGSGFTIETLRYVNAYQKFLELNM 493
Query 570 RKGYSYRDIIEGRFNVKVRYDELLMPEFFGGFSRDIEMHSISQTVDQDLDGSQ-TYAKAL 628
RKG+SY+ I++GR+++ +R+DELLMPEF GG SR++ M ++ QTVDQ S+ YA+AL
Sbjct 494 RKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSGSSRGQYAEAL 553
Query 629 GSQSGIAGVRGDSGRALECFCDEESIVMGILIVTPLPVYTQLLPKHFTYRGLLDHYQPEF 688
GS++GIAGV G + +E FCDEES ++G+L VTP+P+YTQLLPK FTY GLLDHYQPEF
Sbjct 554 GSKTGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQLLPKDFTYNGLLDHYQPEF 613
Query 689 NHIGFQPILYKEVCPLQAYS-DGPDTLSDVFGYNRPWYEYVQKYDQAHGLFR 739
+ IGFQPI YKE+CP+ + D + L+ FGY RPWYEYV KYD AHGLFR
Sbjct 614 DRIGFQPITYKEICPMNIVAGDSANQLNKTFGYQRPWYEYVAKYDSAHGLFR 665
Score = 196 bits (499), Expect = 5e-55, Method: Compositional matrix adjust.
Identities = 93/141 (66%), Positives = 106/141 (75%), Gaps = 6/141 (4%)
Query 1 MANGAFDATLDVNNEIKVNNFDWSHANNLTTQIGRVTPVFCELVPAHGSVRINPRFGLQF 60
MA FDAT D NN I VN+FDWSH NNLTT GR+TPVFCELVPA GS+RINP FGL+
Sbjct 1 MAQNIFDATFDANNRIDVNSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLEL 60
Query 61 MPMVFPIQTRLRARMMFFKYPLRALWDGYRDFIGNFREDLEEPYLDLNTVTRLDAMAKTG 120
MPMVFP+QTR+ AR+ FFK LR++W+ Y DFI NFR+DLEEPY+ L R + M KTG
Sbjct 61 MPMVFPVQTRMFARLNFFKVTLRSMWEDYSDFISNFRDDLEEPYI-LPDSFRFEKMCKTG 119
Query 121 SLGDYLGLPTTLFGKFGTTLS 141
SLGDYLGLPT FGT S
Sbjct 120 SLGDYLGLPT-----FGTKSS 135
> Alpavirinae_Human_gut_21_005_Microviridae_AG016_putative.VP1
Length=742
Score = 388 bits (997), Expect = 9e-125, Method: Compositional matrix adjust.
Identities = 198/380 (52%), Positives = 256/380 (67%), Gaps = 15/380 (4%)
Query 370 YLTSDKPVDLTLGSSPYYNSGSANKDKQIKISAYSFRAYEGIYNAYIRDNRNNPYYVNGQ 429
+ S + ++T P+ S A ++ AY FRAYE +YNAY RD RNNP+ VNG+
Sbjct 302 FFESPEESEITFDDFPF-KSQEAGTKSYPRLLAYRFRAYEAVYNAYYRDIRNNPFVVNGR 360
Query 430 VQYNKWIPTYDGGADQNIYELRYANWEKDFLTTAVQSPQQGT-APLVGIttytetvettS 488
YNKW+PT GGAD+ +Y+L NWE+DFLTTAV +PQQG APLVG+T S
Sbjct 361 PVYNKWLPTMKGGADETMYQLHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTR--S 418
Query 489 DDGTPVTRELSRIALVDEDGKKYQVSF--DSDSEGLKGVSYVELDNEVKLRQPRNLIDVV 546
+DGT ++ + LVDEDG KY +S+ D E L GV Y + + + + ++
Sbjct 419 EDGTLSVQK--QTVLVDEDGSKYGISYKVSEDGERLVGVDYDPVSEKTPVTAINSYAELA 476
Query 547 T------SGISINDLRNVNAYQKFLELNMRKGYSYRDIIEGRFNVKVRYDELLMPEFFGG 600
SG +I LR VNAYQKFLELNMRKG+SY+ I++GR+++ +R+DELLMPEF GG
Sbjct 477 ALATEQGSGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGG 536
Query 601 FSRDIEMHSISQTVDQDLDGSQ-TYAKALGSQSGIAGVRGDSGRALECFCDEESIVMGIL 659
SR++ M ++ QTVDQ SQ YA+ALGS++GIAGV G + +E FCDEES ++G+L
Sbjct 537 ISRELSMRTVEQTVDQQSASSQGQYAEALGSKTGIAGVYGSTSNNIEVFCDEESYIIGLL 596
Query 660 IVTPLPVYTQLLPKHFTYRGLLDHYQPEFNHIGFQPILYKEVCPLQAYSDGPDTLSDVFG 719
VTP+P+Y+QLLPK FTY GLLDHYQPEF+ IGFQPI YKEVCPL + ++ FG
Sbjct 597 TVTPVPIYSQLLPKDFTYNGLLDHYQPEFDRIGFQPITYKEVCPLNFDISNMNDMNKTFG 656
Query 720 YNRPWYEYVQKYDQAHGLFR 739
Y RPWYEYV KYD AHGLFR
Sbjct 657 YQRPWYEYVAKYDNAHGLFR 676
Score = 191 bits (484), Expect = 4e-53, Method: Compositional matrix adjust.
Identities = 85/130 (65%), Positives = 102/130 (78%), Gaps = 1/130 (1%)
Query 1 MANGAFDATLDVNNEIKVNNFDWSHANNLTTQIGRVTPVFCELVPAHGSVRINPRFGLQF 60
MA FDAT D NN I VN+FDWSH NNLTT GR+TPVFCELVPA GS+RINP FGL+
Sbjct 1 MAQNIFDATFDANNRIDVNSFDWSHVNNLTTDFGRITPVFCELVPAKGSLRINPEFGLEL 60
Query 61 MPMVFPIQTRLRARMMFFKYPLRALWDGYRDFIGNFREDLEEPYLDLNTVTRLDAMAKTG 120
MPMVFP+QTR+ AR+ FFK LR++W+ Y DFI NFR+DLEEPY++ + ++ M TG
Sbjct 61 MPMVFPVQTRMFARLNFFKVTLRSMWEDYSDFISNFRDDLEEPYINCSEIS-FPKMFTTG 119
Query 121 SLGDYLGLPT 130
SLGDYLG+PT
Sbjct 120 SLGDYLGVPT 129
> Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1
Length=522
Score = 105 bits (263), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 98/343 (29%), Positives = 143/343 (42%), Gaps = 21/343 (6%)
Query 400 ISAYSFRAYEGIYNAYIRDNRNNPYYVNGQVQYNKWIPTYDGGADQNIYELRYANWEKDF 459
+SA F AY+ IY+ Y RD + +V T G + +R W+ D+
Sbjct 133 VSALPFAAYQKIYDDYYRDEN-----LIDKVDITLSDGTQSGADAIELGTMRKRAWQHDY 187
Query 460 LTTAVQSPQQGTAPLVGIttytetvettSDDGTPVTRELSR---IALVDEDGKKYQVSFD 516
T+A+ Q+G + + T + + + R + IA DG +
Sbjct 188 FTSALPWTQRGPEATIPLGTSAPITWSNNTATATIVRNNTSGNPIAGYTFDGASALYTGG 247
Query 517 SDSEGLKGVSYVELDNEVKLRQPRNLIDVVTSGISINDLRNVNAYQKFLELNMRKGYSYR 576
S + V LD + +L S SINDLR Q++LE N R G Y
Sbjct 248 SGQLIADLPTPVSLDFDNSDHLFADLAGATAS--SINDLRRAFRLQEWLERNARGGARYI 305
Query 577 DIIEGRFNVKVRYDELLMPEFFGGFSRDIEMHSISQTVDQDLDGSQTYAKALGSQSGIAG 636
+II F V+ L PEF GG + I + + QT + T G+ +G G
Sbjct 306 EIIMAHFGVRSSDARLQRPEFLGGSATPITISEVLQT-----SANNTEPTPQGNMAG-HG 359
Query 637 VRGDSGRALECFCDEESIVMGILIVTPLPVYTQLLPKHFTYRGLLDHYQPEFNHIGFQPI 696
V S + C+E ++GI+ V P Y Q +PKHF D+Y P F +IG QPI
Sbjct 360 VSVGSSNYVSYKCEEHGYIIGIMSVMPKTAYQQGIPKHFKKLDKFDYYWPSFANIGEQPI 419
Query 697 LYKEVCPLQAYSDGPDTLSDVFGYNRPWYEYVQKYDQAHGLFR 739
L +E+ Y T ++ FGY + EY HG FR
Sbjct 420 LNEEL-----YHQNNATDTETFGYTPRYAEYKYIPSTVHGEFR 457
Score = 62.4 bits (150), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 37/115 (32%), Positives = 51/115 (44%), Gaps = 2/115 (2%)
Query 17 KVNNFDWSHANNLTTQIGRVTPV-FCELVPAHGSVRINPRFGLQFMPMVFPIQTRLRARM 75
+ N FD SH + +IG + P+ E VP I +F P++ PI +
Sbjct 13 QTNTFDLSHDRKFSGKIGELMPITVMEAVPGD-KFNIKATNLTRFAPLITPIMHQASVYC 71
Query 76 MFFKYPLRALWDGYRDFIGNFREDLEEPYLDLNTVTRLDAMAKTGSLGDYLGLPT 130
FF P R LW + DFI + L +P +T G+L DYLGLPT
Sbjct 72 HFFFVPNRILWSNWEDFISGGEDGLADPTFPTINITTPTNGYAVGALADYLGLPT 126
Score = 27.3 bits (59), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 23/53 (43%), Gaps = 0/53 (0%)
Query 100 LEEPYLDLNTVTRLDAMAKTGSLGDYLGLPTTLFGKFGTTLSVATTGHIFGLK 152
L E N T + T +Y +P+T+ G+F +L G IFG K
Sbjct 420 LNEELYHQNNATDTETFGYTPRYAEYKYIPSTVHGEFRDSLDFWHMGRIFGSK 472
> Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1
Length=505
Score = 102 bits (255), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 98/344 (28%), Positives = 148/344 (43%), Gaps = 41/344 (12%)
Query 400 ISAYSFRAYEGIYNAYIRDNR---NNPY-YVNGQVQYNKWIPTYDGGADQNIYELRYANW 455
+SA F AY+ +YN + RD PY +NG N+ +R +
Sbjct 134 VSALPFAAYQCVYNEWYRDQNLVAEVPYQLINGP------------NPRTNLAVMRLRAF 181
Query 456 EKDFLTTAVQSPQQGTAPLVGIttytetvettSDDGTPVTRELSRIALVDEDGKKYQVSF 515
E D+ T+A+ Q+G A + + T + D P E+ VS
Sbjct 182 EHDYFTSALPFAQKGNAVDIPLGDITLNSDWYLDGSVPKL----------ENSTGGYVSG 231
Query 516 DSDSEGLKGVSYVELDNEVKLRQPRNLIDVVTSGISINDLRNVNAYQKFLELNMRKGYSY 575
+ S+ G V DN+ P ++V ++ +I DLR Q++LEL R G Y
Sbjct 232 NVTSDNATGEINVGSDNDPFAYDPDGSLEVTST--TITDLRRAFKLQEWLELLARGGSRY 289
Query 576 RDIIEGRFNVKVRYDELLMPEFFGGFSRDIEMHSISQTVDQDLDGSQTYAKALGSQSGIA 635
+ I+ F+VK L PE+ G + + + I T +T G+ SG
Sbjct 290 IEQIKTFFDVKSSDQRLQRPEYITGTKQPVIISEILNTT------GETAGLPQGNMSG-H 342
Query 636 GVRGDSGRALECFCDEESIVMGILIVTPLPVYTQLLPKHFTYRGLLDHYQPEFNHIGFQP 695
GV +G + +C+E ++GIL VTP Y Q +PKH+ D Y P+F HIG QP
Sbjct 343 GVAVGTGNVGKYYCEEHGFIIGILSVTPNTAYQQGIPKHYLRTDKYDFYWPQFAHIGEQP 402
Query 696 ILYKEVCPLQAYSDGPDTLSDVFGYNRPWYEYVQKYDQAHGLFR 739
I+ E+ Y+ P T + FGY + EY + G FR
Sbjct 403 IVNNEI-----YAYDP-TGDETFGYTPRYAEYKYMPSRVAGEFR 440
Score = 55.8 bits (133), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 30/119 (25%), Positives = 57/119 (48%), Gaps = 6/119 (5%)
Query 17 KVNNFDWSHANNLTTQIGRVTPVF-CELVPAHGSVRINPRFGLQFMPMVFPIQTRLRARM 75
+ N FD +H ++ ++G + P+ E +P ++ + ++F P++ P+ R M
Sbjct 16 RYNTFDLTHDVKMSGRMGELLPIMNLECIPGD-NITLGADSMVKFAPLLAPVMHRFNVTM 74
Query 76 MFFKYPLRALWDGYRDFIGNFREDLEEPYLDLNTVTRLDAMAKTGSLGDYLGLPTTLFG 134
+F P R +WDG+ DF+ + P +++N+ A DYLG+P G
Sbjct 75 HYFFVPNRIIWDGWEDFMTDVDTPPAFPIIEVNS----GWTAAQQRFADYLGIPPNTAG 129
> Gokush_Human_feces_E_010_Microviridae_AG0120_putative.VP1
Length=538
Score = 98.2 bits (243), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 98/356 (28%), Positives = 142/356 (40%), Gaps = 30/356 (8%)
Query 393 NKDKQIKISAYSFRAYEGIYNAYIRDNRNNPYYVNGQVQYNKWIPTYDGGADQNIYELRY 452
N +K +K++A FRAY I+N + RD +Q + +PT DG + + Y L
Sbjct 133 NVNKALKVNALPFRAYNLIFNEWFRDE---------NLQESLKVPTGDGPDNLSDYSLVR 183
Query 453 ANWEKDFLTTAVQSPQQGTAPLVGIttytetvettSDDGTPVTRELSRIALVDEDGKKYQ 512
D+ T+ + PQ+G + I + G+ + + G
Sbjct 184 RGKRHDYFTSCLPWPQKGPGVEISIGGSATVTFSGGIVGSGSPSFVKSSKITSGQGFLGA 243
Query 513 VSFDSDSEGLK-------GVSYVELD-NEVKLRQPRNLIDVVTSG-ISINDLRNVNAYQK 563
S S SEG S+ L ++ KL P D+ ++ ISINDLR QK
Sbjct 244 GSDISSSEGANVYLKNGINFSFAPLAWDDPKLSLPTATADLSSATPISINDLRQAFQIQK 303
Query 564 FLELNMRKGYSYRDIIEGRFNVKVRYDELLMPEFFGGFSRDIEMHSISQTVDQDLDGSQT 623
E + R G Y +I+ F V L PE+ GG S I ++ + QT + Q
Sbjct 304 LYERDARGGTRYTEILRSHFGVISPDARLQRPEYLGGSSARISINPVQQTSATNETTPQG 363
Query 624 YAKALGSQSGIAGVRGDSGRALECFCDEESIVMGILIVTPLPVYTQLLPKHFTYRGLLDH 683
A GV DS E V G + V Y Q L + ++ +G D
Sbjct 364 NLAAF-------GVVSDSFHGFSKSFVEHGYVFGFVNVRADLTYQQGLNRMWSRQGRFDF 416
Query 684 YQPEFNHIGFQPILYKEVCPLQAYSDGPDTLSDVFGYNRPWYEYVQKYDQAHGLFR 739
Y P H+G Q +L KE+ Y+ G VFGY + EY Q G FR
Sbjct 417 YWPVLAHLGEQAVLNKEI-----YAQGTTDDDKVFGYQERYAEYRYYPGQITGKFR 467
Score = 43.1 bits (100), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 30/120 (25%), Positives = 50/120 (42%), Gaps = 2/120 (2%)
Query 13 NNEIKVNNFDWSHANNLTTQIGRVTPVFCELVPAHGSVRINPRFGLQFMPMVFPIQTRLR 72
+ +I + FD SH T G + P + + V S ++ + + P L
Sbjct 17 STQIPRSVFDRSHGYKTTFNSGFLVPFYVDEVLPGDSFKLTATLFARLATPIVPFMDNLY 76
Query 73 ARMMFFKYPLRALWDGYRDFIGNFREDLEEPYLDLNTVTRLDAMAKTGSLGDYLGLPTTL 132
FF P R +WD ++ F G + + + TV+ + +T L DY GLPT +
Sbjct 77 LETFFFFVPNRLVWDNWQKFNGEQKNPSDPTDFLIPTVSGTNVQNQT--LWDYFGLPTNV 134
> Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1
Length=508
Score = 97.4 bits (241), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 94/349 (27%), Positives = 143/349 (41%), Gaps = 39/349 (11%)
Query 400 ISAYSFRAYEGIYNAYIRD-NRNNPYYVNGQVQYNKWIPTYDGGADQNIYELRYANWEKD 458
+SA AY+ IYN Y RD N P + D G + + LR WE D
Sbjct 135 VSALPMAAYQCIYNEYYRDQNLQTPV----DYKLTDGNNNTDAGDRERLTTLRQRAWEHD 190
Query 459 FLTTAVQSPQQGTA---PLVGIttytetvettSDDGTPVTRELSRIALVDEDGKKYQVSF 515
+ T ++ Q+G A P+ I + + G R S L+D
Sbjct 191 YFTASLPFAQKGAAVDIPIGTIDKDVAVNFNSLEFGATTVRYNSADGLID---------- 240
Query 516 DSDSEGLKGVSYVELDNEVKLRQPRNLIDVVTSGISINDLRNVNAYQKFLELNMRKGYSY 575
+ G + V D + + ID+ + +INDLR Q++LE N R G Y
Sbjct 241 ------IGGSNAVNADGTPTMIAKTSTIDIEPT--TINDLRRAFKLQEWLEKNARGGTRY 292
Query 576 RDIIEGRFNVKVRYDELLMPEFFGGFSRDIEMHSISQTVDQDLDGSQTYAKALGSQSGIA 635
+ I F V+ L PE+ G + + + T QD Q G+ +G
Sbjct 293 IENILTHFGVRSSDKRLQRPEYITGVKSPVVVSEVLNTTGQDGGLPQ------GNMAG-H 345
Query 636 GVRGDSGRALECFCDEESIVMGILIVTPLPVYTQLLPKHFTYRGLLDHYQPEFNHIGFQP 695
G+ SG++ +C+E ++GI+ V P Y Q +P+ F LD++ P F +IG Q
Sbjct 346 GISVTSGKSGSYYCEEHGYIIGIMSVMPKTAYQQGIPRTFLKTDSLDYFWPTFANIGEQE 405
Query 696 ILYKEVCPLQAYSDGPDTLSDVFGYNRPWYEYVQKYDQAHGLFRRPEQY 744
+ +E L AY+ + D FGY + EY + G FR Y
Sbjct 406 VAKQE---LYAYTANAN---DTFGYVPRYAEYKYMPSRVAGEFRTSLNY 448
Score = 63.9 bits (154), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 39/130 (30%), Positives = 60/130 (46%), Gaps = 5/130 (4%)
Query 14 NEIKVNNFDWSHANNLTTQIGRVTPVFC-ELVPAHGSVRINPRFGLQFMPMVFPIQTRLR 72
N+ K N FD +H L+T +G++TP+ E VP ++ ++F PM+ P+ R+
Sbjct 12 NKPKKNVFDLTHDVKLSTNMGQLTPILTLECVPGD-KFDLSCESLIRFAPMIAPVMHRMD 70
Query 73 ARMMFFKYPLRALWDGYRDFIGNFREDLEEPYLDLNTVTRLDAMAKTGSLGDYLGLPTTL 132
M +F P R LW + FI + PY+ T D A DY+G+P
Sbjct 71 VTMHYFFVPNRILWSNWEKFITEHNSEHVAPYM---AYTNGDYTAMQKKFMDYIGIPPVP 127
Query 133 FGKFGTTLSV 142
G T +S
Sbjct 128 AGGVSTNVSA 137
> Gokush_Bourget_915_Microviridae_AG0177_putative.VP1
Length=549
Score = 96.3 bits (238), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 101/384 (26%), Positives = 141/384 (37%), Gaps = 60/384 (16%)
Query 390 GSANKDKQIKISAYSFRAYEGIYNAYIRDNRNNPYYVNGQVQYNKWIPTYDG--GADQNI 447
G + SA RAY IYN + RD +Q ++ + DG +
Sbjct 136 GQVTAGATVSHSALPTRAYNLIYNQWFRDE---------NLQNSRVVDKGDGPDATPSTV 186
Query 448 YELRYANWEKDFLTTAVQSPQQG----------TAPLVGIttytetvettSDDGTPVTRE 497
Y L+ D+ T+++ PQ+G +AP+ G D T +
Sbjct 187 YALQRRGKRHDYFTSSLPWPQKGGTAVSLPLGTSAPVYGTGKTLGLF----DGVTNLGLH 242
Query 498 LSRIALVDEDGKKYQVSFDSDSEGLKGVSYVELDNEVKLRQPRNLIDVVTSGIS------ 551
+ Y + S G+ GV L VVTSG+S
Sbjct 243 ADTTNTLRNASSSYNTNIGSIPTGVGGVISKNLG-------------VVTSGVSGLYADL 289
Query 552 -------INDLRNVNAYQKFLELNMRKGYSYRDIIEGRFNVKVRYDELLMPEFFGGFSRD 604
IN LR QK LE + R G Y +I+ F V L PE+ GG S
Sbjct 290 SAATSATINQLRQSFQIQKLLERDARGGTRYTEILRSHFGVTSPDARLQRPEYLGGGSTL 349
Query 605 IEMHSISQTVDQDLDGSQTYAKALGSQSGIAGVRGDSGRALECFCDEESIVMGILIVTPL 664
I + I+QT + G T L + GV E V+G++ V
Sbjct 350 INISPIAQTTGTGISGQTTPQGNLAAM----GVYHAHNHGFTQSFVEHGYVIGVIAVRAD 405
Query 665 PVYTQLLPKHFTYRGLLDHYQPEFNHIGFQPILYKEVCPLQAYSDGPDTLSDVFGYNRPW 724
Y Q L +H++ D+Y P F +G Q IL KE+ Y G + S+VFGY W
Sbjct 406 LTYQQGLRRHWSRSTRYDYYFPAFAMLGEQAILNKEI-----YVTGGSSDSNVFGYQERW 460
Query 725 YEYVQKYDQAHGLFRRPEQYEIIP 748
EY + GLFR I P
Sbjct 461 AEYRYNPSEITGLFRSTAAGTIDP 484
Score = 42.4 bits (98), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/108 (27%), Positives = 42/108 (39%), Gaps = 0/108 (0%)
Query 34 GRVTPVFCELVPAHGSVRINPRFGLQFMPMVFPIQTRLRARMMFFKYPLRALWDGYRDFI 93
G + P+ CE V + +N + +FP+ L FF P R +W + F+
Sbjct 38 GLLVPIMCEEVLPGDTFNVNVTMFGRLATPIFPVMDNLHLDSFFFFVPNRLVWTNWVKFM 97
Query 94 GNFREDLEEPYLDLNTVTRLDAMAKTGSLGDYLGLPTTLFGKFGTTLS 141
G + + GSL DYLGLPT G T+S
Sbjct 98 GEQDNPADSISYTIPQQVSPAGGYAIGSLQDYLGLPTVGQVTAGATVS 145
> Gokush_Human_feces_C_031_Microviridae_AG0433_putative.VP1
Length=529
Score = 96.3 bits (238), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 100/354 (28%), Positives = 145/354 (41%), Gaps = 33/354 (9%)
Query 392 ANKDKQIKISAYSFRAYEGIYNAYIRDNRNNPYYVNGQVQYNKWIPTYDGGADQNIYELR 451
N + +K++A FRAY I+N + RD +Q + +PT DG + + Y L
Sbjct 132 TNVSEALKVNALPFRAYNLIFNEWFRDE---------NLQESLKVPTGDGPDNLSDYSLV 182
Query 452 YANWEKDFLTTAVQSPQQGTAPLVGIttytetvettSDDGTPVTRELSRIALVDEDGKKY 511
D+ T+ + PQ+G P V I+ + S +GTP T ++S + G +
Sbjct 183 RRGKRHDYFTSCLPWPQKG--PGVEISIGGSANVSFSGNGTP-TFDISSSNVSLPAGGLF 239
Query 512 QVSFDSDSEGLKGVSYVELDNEVKLRQPRNLIDVVTSG------ISINDLRNVNAYQKFL 565
D+ GV N +L ++ V T+ ISINDLR QK
Sbjct 240 VA---GDTNTSVGVYRGGTANTGRLFSWKDSGLVATADLSTATPISINDLRQAFQIQKLY 296
Query 566 ELNMRKGYSYRDIIEGRFNVKVRYDELLMPEFFGGFSRDIEMHSISQTVDQDLDGSQTYA 625
E + R G Y +I+ F V L PE+ GG S I ++ + QT + Q
Sbjct 297 ERDARGGTRYTEILRSHFGVISPDARLQRPEYLGGSSARISINPVQQTSATNDTTPQGNL 356
Query 626 KALGSQSGIAGVRGDSGRALECFCDEESIVMGILIVTPLPVYTQLLPKHFTYRGLLDHYQ 685
A GV DS E V G + V Y Q L + ++ +G D Y
Sbjct 357 AAF-------GVVSDSFHGFSKSFVEHGYVFGFVNVRADLTYQQGLNRMWSRQGRFDFYW 409
Query 686 PEFNHIGFQPILYKEVCPLQAYSDGPDTLSDVFGYNRPWYEYVQKYDQAHGLFR 739
P H+G Q +L KE+ Y+ G VFGY + EY Q G FR
Sbjct 410 PVLAHLGEQAVLNKEI-----YAQGTADDDKVFGYQERYAEYRYYPGQITGKFR 458
Score = 43.9 bits (102), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 30/120 (25%), Positives = 50/120 (42%), Gaps = 2/120 (2%)
Query 13 NNEIKVNNFDWSHANNLTTQIGRVTPVFCELVPAHGSVRINPRFGLQFMPMVFPIQTRLR 72
+ +I + FD SH T G + P + + V S ++ + + P L
Sbjct 17 STQIPRSVFDRSHGYKTTFNSGYLVPFYVDEVLPGDSFKLTATLFARLATPIVPFMDNLY 76
Query 73 ARMMFFKYPLRALWDGYRDFIGNFREDLEEPYLDLNTVTRLDAMAKTGSLGDYLGLPTTL 132
FF P R +WD ++ F G + + + TV+ + +T L DY GLPT +
Sbjct 77 LETFFFFVPNRLVWDNWQKFNGEQKNPTDSTDFLIPTVSGTNVQNQT--LWDYFGLPTNV 134
> Gokush_gi|12085136|ref|NP_073538.1|_major_capsid_protein_[Bdellovibrio_phage_phiMH2K]
Length=533
Score = 94.4 bits (233), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 97/359 (27%), Positives = 151/359 (42%), Gaps = 26/359 (7%)
Query 384 SPYYNSGSANKDKQIKISAYSFRAYEGIYNAYIRDNRNNPYYVNGQVQYNKWIPTYDGGA 443
S Y + G + ++I+A FRAY IYN + RD + G++ +P DG
Sbjct 125 SIYDHFGIPTQVANLEINALPFRAYNLIYNDWFRDQN-----LIGKIA----VPKGDGPD 175
Query 444 DQNIYELRYANWEKDFLTTAVQSPQQGTAPLVGIttytetvettSDDGTPVTR--ELSRI 501
+ Y+L A D+ T+A+ PQ+G A + I + P +
Sbjct 176 NHADYQLLKAAKPHDYFTSALPWPQKGMAVEMPIGNSAPITYVPNAGNGPYPHFNWVQTP 235
Query 502 ALVDEDGKKYQVSFDSDSEGLKGVSYVELDNEVKLRQPRNLIDVVT-SGISINDLRNVNA 560
+G QV+F G K +S D Q + D+ + + +IN LR
Sbjct 236 GGPGNNGALSQVTFG----GQKAISAAGNDPIGYDPQGTLIADLSSATAATINQLRQAMM 291
Query 561 YQKFLELNMRKGYSYRDIIEGRFNVKVRYDELLMPEFFGGFSRDIEMHSISQTVDQDLDG 620
Q LEL+ R G Y +I++ FNV L PE+ G + D++ + + QT D
Sbjct 292 MQSLLELDARGGTRYVEILKSHFNVISLDFRLQRPEYLSGGTIDLQQNPVPQTSSSTTDS 351
Query 621 SQTYAKALGSQSGIAGVRGDSGRALECFCDEESIVMGILIVTPLPVYTQLLPKHFTYRGL 680
Q A + S G S + F E V+G + Y Q L K ++ +
Sbjct 352 PQGNLAAFSTASEFGNKIGFS----KSFV-EHGYVLGFIRARGQVTYQQGLHKMWSRQTR 406
Query 681 LDHYQPEFNHIGFQPILYKEVCPLQAYSDGPDTLSDVFGYNRPWYEYVQKYDQAHGLFR 739
D + P+F +G Q IL KE+ Y+ G T S++FGY + EY + + G FR
Sbjct 407 WDFFWPKFQELGEQAILNKEI-----YAQGNATDSEIFGYQERYGEYRFRPSEIKGQFR 460
Score = 41.6 bits (96), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/114 (22%), Positives = 50/114 (44%), Gaps = 0/114 (0%)
Query 19 NNFDWSHANNLTTQIGRVTPVFCELVPAHGSVRINPRFGLQFMPMVFPIQTRLRARMMFF 78
+ F+ S T + +TP+F + + ++ +N + ++ V P+ R+ FF
Sbjct 23 SKFNRSFGTKDTFKFDDLTPIFIDEILPGDTINMNTKTFIRLATQVVPVMDRMMLDFYFF 82
Query 79 KYPLRALWDGYRDFIGNFREDLEEPYLDLNTVTRLDAMAKTGSLGDYLGLPTTL 132
P R +WD + F G + + T+T + S+ D+ G+PT +
Sbjct 83 FVPCRLVWDNWEKFNGAQDNPSDSTDYLIPTITAPAGGFENMSIYDHFGIPTQV 136
Lambda K H a alpha
0.319 0.137 0.411 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 70204668