bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
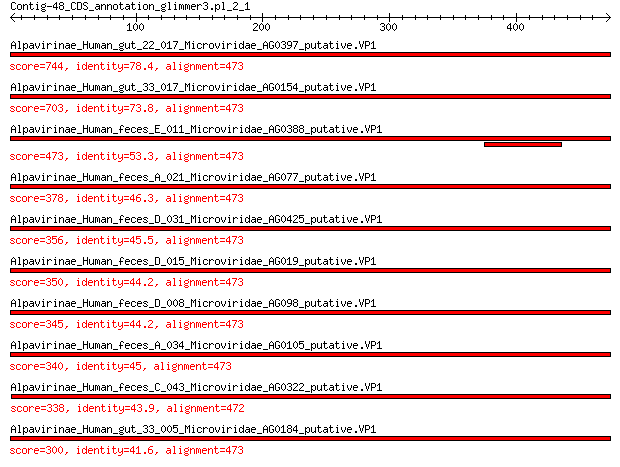
Query= Contig-48_CDS_annotation_glimmer3.pl_2_1
Length=473
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1 744 0.0
Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1 703 0.0
Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1 473 2e-162
Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1 378 6e-126
Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1 356 3e-117
Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1 350 1e-115
Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1 345 4e-113
Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1 340 2e-111
Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1 338 1e-110
Alpavirinae_Human_gut_33_005_Microviridae_AG0184_putative.VP1 300 9e-96
> Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1
Length=607
Score = 744 bits (1921), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 371/474 (78%), Positives = 405/474 (85%), Gaps = 8/474 (2%)
Query 1 LGFSGADLAYKLLSYLGYGNFIPSPPSNSTRWWSTSLKKEASPTGYTQQYIQNNYVNLFP 60
GFS ADLA+KL SYLGYGN S S+S RWWSTSLK S YTQQY+Q++YVNLFP
Sbjct 141 FGFSRADLAFKLFSYLGYGNVWSSEFSSSNRWWSTSLKGGGS---YTQQYVQDSYVNLFP 197
Query 61 LLAYQKIYQDFFRWSQWERANPSSYNVDYYSGVSSSLVTVLPDYTSDYWKSDTMFDLKYC 120
LLAYQKIYQDFFRWSQWE +NPSSYNVDY++GVS LV+ LP+ ++ YWKS TMFDLKYC
Sbjct 198 LLAYQKIYQDFFRWSQWENSNPSSYNVDYFTGVSPLLVSSLPEASNAYWKSPTMFDLKYC 257
Query 121 NWNKDMLMGILPDSQFGECCYKSIFETPGGDLKAGFRTTDGKFISAVTNAPLTTENSSS- 179
NWNKDMLMG+LP+SQFG+ I + D+ G G S V A + N++S
Sbjct 258 NWNKDMLMGVLPNSQFGDVAVLDIDNSGKPDVVLGL----GNANSTVGVASYVSSNTASI 313
Query 180 GLSTPGVTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSGDSDYREQIR 239
+S +T+ + S L DL++L+SQF+VLALRQAEALQRWKEISQSGDSDYREQIR
Sbjct 314 PFFALKASSANTLPVGSTLRVDLASLKSQFTVLALRQAEALQRWKEISQSGDSDYREQIR 373
Query 240 KHFGVNLPQSLSNLCTYIGGISRNLDISEVVNNNLAAEGDTAVIAGKGVGAGNGSFTYTT 299
KHFGVNLPQ+LSN+CTYIGGISRNLDISEVVNNNLAAEGDTAVIAGKGVGAGNGSFTYTT
Sbjct 374 KHFGVNLPQALSNMCTYIGGISRNLDISEVVNNNLAAEGDTAVIAGKGVGAGNGSFTYTT 433
Query 300 DEHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGMEVLPMTQIFNSPKAS 359
+EHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGME LPMTQIFNSPKAS
Sbjct 434 NEHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGMETLPMTQIFNSPKAS 493
Query 360 IVNLFNAGYNPRYFNWKTKLDVVNGAFTTTLKSWVSPVTESLLSGWFGFGYSQDDVNKDT 419
IVNLFNAGYNPRYFNWKTKLDV+NGAFTTTLKSWVSPVTESLLSGWFGFGYS+ DVN
Sbjct 494 IVNLFNAGYNPRYFNWKTKLDVINGAFTTTLKSWVSPVTESLLSGWFGFGYSEGDVNSQN 553
Query 420 KVVLNYKFFKVNPSVLDPIFGVNADSTWDTDQLLVNSYIGCYVARNLSRDGVPY 473
KVVLNYKFFKVNPSVLDPIFGV ADSTWD+DQLLVNSYIGCYVARNLSRDGVPY
Sbjct 554 KVVLNYKFFKVNPSVLDPIFGVAADSTWDSDQLLVNSYIGCYVARNLSRDGVPY 607
> Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1
Length=614
Score = 703 bits (1815), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 349/473 (74%), Positives = 388/473 (82%), Gaps = 5/473 (1%)
Query 1 LGFSGADLAYKLLSYLGYGNFIPSPPSNSTRWWSTSLKKEASPTGYTQQYIQNNYVNLFP 60
GFS ADL++KLLSYLGYGN I +PPS RWWSTSLK YTQQYIQN VN+FP
Sbjct 147 FGFSRADLSFKLLSYLGYGNLIETPPSLGNRWWSTSLKNTDDGANYTQQYIQNTIVNIFP 206
Query 61 LLAYQKIYQDFFRWSQWERANPSSYNVDYYSGVSSSLVTVLPDYTSDYWKSDTMFDLKYC 120
LL YQKIYQDFFRW QWE++NPSSYNVDY+SG S S+V+ LP +S YWKSDTMFDLKYC
Sbjct 207 LLTYQKIYQDFFRWPQWEKSNPSSYNVDYFSGSSPSIVSDLPAASSAYWKSDTMFDLKYC 266
Query 121 NWNKDMLMGILPDSQFGECCYKSIFETPGGDLKAGFRTTDGKFISAVTNAPLTTENSSSG 180
NWNKDMLMG+LP+SQFG+ I + D+ G D + + +T++++
Sbjct 267 NWNKDMLMGVLPNSQFGDVAVLDISSSGDSDVVLG---VDPHKSTLGIGSAITSKSAVVP 323
Query 181 LSTPGVTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSGDSDYREQIRK 240
L ++ + V++ S L DLS+++SQF+VLALRQAEALQRWKEISQSGDSDYREQI K
Sbjct 324 LFALDASTSNPVSVGSKLHLDLSSIKSQFNVLALRQAEALQRWKEISQSGDSDYREQILK 383
Query 241 HFGVNLPQSLSNLCTYIGGISRNLDISEVVNNNLAAEGDTAVIAGKGVGAGNGSFTYTTD 300
HFGV LPQ+LSNLCTYIGGISRNLDISEVVNNNLAAE DTAVIAGKGVG GNGSFTYTT+
Sbjct 384 HFGVKLPQALSNLCTYIGGISRNLDISEVVNNNLAAEEDTAVIAGKGVGTGNGSFTYTTN 443
Query 301 EHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGMEVLPMTQIFNSPKASI 360
EHCV+M IYHAVPLLDYT+TGQDGQLLVTDAESLPIPEFDNIG+EVLPM QIFNS A+
Sbjct 444 EHCVIMGIYHAVPLLDYTLTGQDGQLLVTDAESLPIPEFDNIGLEVLPMAQIFNSSLATA 503
Query 361 VNLFNAGYNPRYFNWKTKLDVVNGAFTTTLKSWVSPVTESLLSGWFGFGYSQDDVNKDTK 420
NLFNAGYNPRYFNWKTKLDV+NGAFTTTLKSWVSPV+ESLLSGW FG S D TK
Sbjct 504 FNLFNAGYNPRYFNWKTKLDVINGAFTTTLKSWVSPVSESLLSGWARFGAS--DSKTGTK 561
Query 421 VVLNYKFFKVNPSVLDPIFGVNADSTWDTDQLLVNSYIGCYVARNLSRDGVPY 473
VLNYKFFKVNPSVLDPIFGV ADSTWDTDQLLVNSYIGCYV RNLSRDGVPY
Sbjct 562 AVLNYKFFKVNPSVLDPIFGVKADSTWDTDQLLVNSYIGCYVVRNLSRDGVPY 614
> Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1
Length=622
Score = 473 bits (1216), Expect = 2e-162, Method: Compositional matrix adjust.
Identities = 252/486 (52%), Positives = 335/486 (69%), Gaps = 26/486 (5%)
Query 1 LGFSGADLAYKLLSYLGYGNFIPSPPSN----STRWWSTSLKKEASPTGYTQQYIQNNYV 56
G++ D+ KLL YL YGNF+ SN S RWW+TS P GY+Q+Y+ NN V
Sbjct 150 FGYNRGDVDSKLLFYLNYGNFVNPSLSNVGSPSNRWWNTSFSSSKVP-GYSQKYLNNNAV 208
Query 57 NLFPLLAYQKIYQDFFRWSQWERANPSSYNVDYYSG----VSSSLVTVLPDYTSDYWKSD 112
N+FPLLAYQKIYQDFFRWSQWE A+P+SYNVDYY+G SS ++ ++ YWK D
Sbjct 209 NIFPLLAYQKIYQDFFRWSQWENADPTSYNVDYYNGSGNLFGSSGLSGSVTASNSYWKRD 268
Query 113 TMFDLKYCNWNKDMLMGILPDSQFGECCYKSIFETPGGDLKAGFRTTDGKFISAVTNAPL 172
MF L+YCNWNKDM G+LP+SQFG+ ++ ++ G + GF + F A +
Sbjct 269 NMFSLRYCNWNKDMFTGLLPNSQFGDVAVVNLGDSGSGTIPVGFLSDTEVFTQAFNATSM 328
Query 173 TTENSSSGLSTPGVTSGSTVALKSPLISDLS-ALQSQFSVLALRQAEALQRWKEISQSGD 231
+T + +S + G++ + V+ + +++ ++ A + FS+LALRQAEALQ+WKEI+QS D
Sbjct 329 STVSDTSPM---GISGSTPVSARQSMVARINNADVASFSILALRQAEALQKWKEITQSVD 385
Query 232 SDYREQIRKHFGVNLPQSLSNLCTYIGGISRNLDISEVVNNNLAAEGDTAVIAGKGVGAG 291
++YR+QI+ HFG+N P S+S++ YIGG++RNLDISEVVNNNL +G AVI GKGVG+G
Sbjct 386 TNYRDQIKAHFGINTPASMSHMAQYIGGVARNLDISEVVNNNLKDDGSEAVIYGKGVGSG 445
Query 292 NGSFTYTT-DEHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGMEVLPMT 350
+G Y T ++C++MCIYHA+PLLDY ITGQD QLL T E LPIPEFDNIGME +P T
Sbjct 446 SGKMRYHTGSQYCIIMCIYHAMPLLDYAITGQDPQLLCTSVEDLPIPEFDNIGMEAVPAT 505
Query 351 QIFNSP--KASIVNLFNAGYNPRYFNWKTKLDVVNGAFTTTLKSWVSPVTESLLSGWFGF 408
+FNS + VN F GYNPRY+ WK+K+D V+GAFTTTLK WV+P+ + L WF
Sbjct 506 TLFNSVLFDGTAVNDF-LGYNPRYWPWKSKIDRVHGAFTTTLKDWVAPIDDDYLHNWFN- 563
Query 409 GYSQDDVNKDTK-VVLNYKFFKVNPSVLDPIFGVNADSTWDTDQLLVNSYIGCYVARNLS 467
+KD K +++ FFKVNP+ LD IF V ADS W+TDQLL+N + C V R LS
Sbjct 564 -------SKDGKSASISWPFFKVNPNTLDSIFAVVADSIWETDQLLINCDVSCKVVRPLS 616
Query 468 RDGVPY 473
+DG+PY
Sbjct 617 QDGMPY 622
Score = 28.1 bits (61), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 35/64 (55%), Gaps = 9/64 (14%)
Query 375 WKTKLDVVNGAFTTTLKSWVSPV--TESLLSGWFG--FGYSQDDVNKDTKVVLNYKFFKV 430
W T LD+ N + S SP+ T S+ SG FG FGY++ DV+ LNY F V
Sbjct 117 WCTLLDLSNAVY---FSSGSSPLGSTVSVPSG-FGNIFGYNRGDVDSKLLFYLNYGNF-V 171
Query 431 NPSV 434
NPS+
Sbjct 172 NPSL 175
> Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1
Length=622
Score = 378 bits (971), Expect = 6e-126, Method: Compositional matrix adjust.
Identities = 219/492 (45%), Positives = 283/492 (58%), Gaps = 32/492 (7%)
Query 1 LGFSGADLAYKLLSYLGYGNFI-PSPPSNSTRWWSTSLKKEASPTGYTQQYIQNNYVNLF 59
GFS ADL+ KLL L YGNFI P T + S K A + Y+ N VN+
Sbjct 144 FGFSRADLSAKLLQMLKYGNFINPEHSGLDTPMFGYSTVKLAQFS-----YLWNQAVNVL 198
Query 60 PLLAYQKIYQDFFRWSQWERANPSSYNVDYYSGVSS-SLVTVLPDYTSDYWKSDTMFDLK 118
P+ YQKIY D+FR+ QWE+ P +YN DYY+G T +PD YW + T FDL+
Sbjct 199 PIFCYQKIYSDYFRFQQWEKPVPYTYNADYYAGGDIFPNPTTMPD---AYWNNYTPFDLR 255
Query 119 YCNWNKDMLMGILPDSQFGECCYKSIFETPGGDLKA--GFRTTDGKFISAVTNAPLTTEN 176
YCNWN+D+ G LP+ QFG + + + A F T + AVT A L +
Sbjct 256 YCNWNRDLYTGFLPNQQFGNVSVVDMTVSSDEIMAAPVTFGATGNRV--AVTKAALASST 313
Query 177 SSSGLSTPGVTSGSTVALKSPLISDLSA--LQSQFSVLALRQAEALQRWKEISQSGDSDY 234
SS+G+ T SG TV + + L + + L FS+LALRQAE LQ+WKEI+ SGD DY
Sbjct 314 SSTGIGTVSTPSGGTVPVGTSLYAQVQQRDLTGAFSILALRQAEFLQKWKEIALSGDQDY 373
Query 235 REQIRKHFGVNLPQSLSNLCTYIGGISRNLDISEVVNNNLAAEGDT------AVIAGKGV 288
R QI KHFGV LP LS + YIGG +DISEVVN NL + + A+IAGKGV
Sbjct 374 RSQIEKHFGVKLPAELSYMSQYIGGQFAQMDISEVVNQNLTDQAGSDAAQYPALIAGKGV 433
Query 289 GAGNGSFTYTTDEHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGMEVLP 348
+G+G+ YT +H ++M IYHAVPLLDY TGQD LL+T AE IPEFD IGM+ LP
Sbjct 434 NSGDGNVNYTARQHGIIMGIYHAVPLLDYERTGQDQDLLITSAEEWAIPEFDAIGMQTLP 493
Query 349 MTQIFNSPKASIVNLFN-------AGYNPRYFNWKTKLDVVNGAFTTTLKSWVSPVTESL 401
+ +FNS K S + F GY PRY NWKT +D + GAF ++ K+WV+P+
Sbjct 494 LGTLFNSNKVSGDSDFRLHGAAYPIGYVPRYVNWKTDIDEIFGAFRSSEKTWVAPIDADF 553
Query 402 LSGWFGFGYSQDDVNKDTKVVLNYKFFKVNPSVLDPIFGVNADSTWDTDQLLVNSYIGCY 461
++ W + D + + NY +FKVNP++LD IF V ADST DTD +
Sbjct 554 ITNWVK---NVADNASAVQSLFNYNWFKVNPAILDDIFAVKADSTMDTDTFKTRMMMSIK 610
Query 462 VARNLSRDGVPY 473
RNL G+PY
Sbjct 611 CVRNLDYSGMPY 622
> Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1
Length=614
Score = 356 bits (913), Expect = 3e-117, Method: Compositional matrix adjust.
Identities = 215/501 (43%), Positives = 281/501 (56%), Gaps = 57/501 (11%)
Query 1 LGFSGADLAYKLLSYLGYGNFIPSPPS--NSTRWWSTSL---KKEASPTGYTQQYIQNNY 55
LGF + KLL YL YGNF+ S S N + S SL E + TGYT +I
Sbjct 143 LGFDAITQSAKLLMYLRYGNFLSSVVSDKNKSLGLSGSLDLRNSETASTGYTSMHI---- 198
Query 56 VNLFPLLAYQKIYQDFFRWSQWERANPSSYNVDYYSG--VSSSLVTVLPDYTSDYWKSDT 113
PL AYQK Y DFFR++QWE+ P +YN D+YSG V +SL T+ D Y+ D
Sbjct 199 ---LPLAAYQKAYADFFRFTQWEKNQPYTYNFDWYSGGNVLASLTTL--DLAKKYYSDDN 253
Query 114 MFDLKYCNWNKDMLMGILPDSQFGECCYKSIFETPG--------------GDLKAGFRTT 159
+F L+Y NW KDM MG++PDSQ G+ SI + G G L+AG
Sbjct 254 LFTLRYANWPKDMFMGVMPDSQLGDV---SIVDASGSEGTFPVGLLDVNDGTLRAGLLAR 310
Query 160 DGKFISAVTNAPLTTENSSSGLSTPGVTSGSTVALKSPLISDLSALQSQFSVLALRQAEA 219
G + ++ + T ++ S +T GV + + L S FS+L LR AEA
Sbjct 311 SGSAPAEKSSLEMQTSSALSANTTYGVYAQRA-----------AGLASSFSILQLRMAEA 359
Query 220 LQRWKEISQSGDSDYREQIRKHFGVNLPQSLSNLCTYIGGISRNLDISEVVNNNLAAEGD 279
+Q+++E+SQ D D R QI HFGV+L LS+ C Y+GG S N+D+SEVVN N+ + +
Sbjct 360 VQKYREVSQFADQDARGQIMAHFGVSLSPVLSDKCVYLGGSSSNIDLSEVVNTNITGD-N 418
Query 280 TAVIAGKGVGAGNGSFTYTTDEHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEF 339
A IAGKGVG G GSF+ DE+ +++ IYH VPLLDY ITGQ LL T+ LP PEF
Sbjct 419 VAEIAGKGVGTGQGSFSGQFDEYGIIIGIYHNVPLLDYVITGQPQNLLYTNTADLPFPEF 478
Query 340 DNIGMEVLPMTQIFNSPKASIVN-----LFNAGYNPRYFNWKTKLDVVNGAFTTTLKSWV 394
D+IGM+ + + NS S + + GY PR+F+ KT+ D V GAF +TLK+WV
Sbjct 479 DSIGMQTIQFGRFVNSKSVSWTSGVDYRVQTMGYLPRFFDVKTRYDEVLGAFRSTLKNWV 538
Query 395 SPVTESLLSGWFGFGYSQDDVNKDTKVV--LNYKFFKVNPSVLDPIFGVNADSTWDTDQL 452
+P+ S +S W Q V K+ LNY FFKVNP VLD IF V DST DTDQ
Sbjct 539 APLDPSYVSKWL-----QSSVTSSGKLALNLNYGFFKVNPRVLDSIFNVKCDSTIDTDQF 593
Query 453 LVNSYIGCYVARNLSRDGVPY 473
L Y+ RN DG+PY
Sbjct 594 LTALYMDIKAVRNFDYDGMPY 614
> Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1
Length=584
Score = 350 bits (899), Expect = 1e-115, Method: Compositional matrix adjust.
Identities = 209/484 (43%), Positives = 285/484 (59%), Gaps = 45/484 (9%)
Query 1 LGFSGADLAYKLLSYLGYGNFIPSPPSNSTRWWSTSLKKEASPTGYTQQYIQNNYVNLFP 60
GF+ ADLAYKL+ YL YGN ++ +R + TS+ + + Y Q N+ +++FP
Sbjct 135 FGFNRADLAYKLIQYLRYGNVRTGVGTSGSRNYGTSV--DVKDSSYNQNRAFNHALSVFP 192
Query 61 LLAYQKIYQDFFRWSQWERANPSSYNVDYYSGVSSSLVTVLPDYTSD---YWKSDTMFDL 117
+LAY+K QD+FR +QW+ + P +N+DYY G ++ T+LP S Y++ DT FDL
Sbjct 193 ILAYKKFCQDYFRLTQWQVSAPYLWNIDYYDGKGAT--TILPADLSKSVTYFEHDTFFDL 250
Query 118 KYCNWNKDMLMGILPDSQFGECCYKSIFETPGGDLKAGFRTTDGKFISAVT-NAPLTTEN 176
+YCNWNKDM G LPD+Q+G+ I + TT I+A +P+ N
Sbjct 251 EYCNWNKDMFFGSLPDAQYGDTSVVDI----------SYGTTGAPVITAQNLQSPV---N 297
Query 177 SSSGLSTPGVTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSGDSDYRE 236
SS+ + T S + + L D VLALR+ EALQR++EIS +YR
Sbjct 298 SSTAIGTSDKFSTQLIEAGTNLTLD---------VLALRRGEALQRFREISLCTPLNYRS 348
Query 237 QIRKHFGVNLPQSLSNLCTYIGGISRNLDISEVVNNNLAAEGDTAVIAGKGVGAGNGSFT 296
QI+ HFGV++ LS + TYIGG + +LDISEVVN N+ E + A+IAGKG+G G G+
Sbjct 349 QIKAHFGVDVGSELSGMSTYIGGEASSLDISEVVNTNIT-ESNEALIAGKGIGTGQGNEE 407
Query 297 YTTDEHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGMEVLPMTQIFNSP 356
+ + V+MCIYH+VPLLDY I+ D QL + S P+PE D IG+E + + N+P
Sbjct 408 FYAKDWGVLMCIYHSVPLLDYVISAPDPQLFASMNTSFPVPELDAIGLEPITVAYYSNNP 467
Query 357 KA--SIVNLFNA-----GYNPRYFNWKTKLDVVNGAFTTTLKSWVSPVTESLLSGWFGFG 409
S + +A GY PRY+ WKT +D V GAFTTT K WV+P+T L S
Sbjct 468 IELPSTGGITDAPTTTVGYLPRYYAWKTSIDYVLGAFTTTEKEWVAPITPELWSNML--- 524
Query 410 YSQDDVNKDTKVVLNYKFFKVNPSVLDPIFGVNADSTWDTDQLLVNSYIGCYVARNLSRD 469
+ K T + NY FFKVNPS+LDPIF VNADS WDTD L+N+ VARNL D
Sbjct 525 --KPLGTKGTGI--NYNFFKVNPSILDPIFAVNADSYWDTDTFLINAAFDIRVARNLDYD 580
Query 470 GVPY 473
G+PY
Sbjct 581 GMPY 584
> Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1
Length=618
Score = 345 bits (885), Expect = 4e-113, Method: Compositional matrix adjust.
Identities = 209/490 (43%), Positives = 274/490 (56%), Gaps = 35/490 (7%)
Query 1 LGFSGADLAYKLLSYLGYGNFIPS--PPSNSTRWWSTSL---KKEASPTGYTQQYIQNNY 55
LGF + KLL YL YGNF+ S P + S+SL E TGYT
Sbjct 147 LGFDSITQSAKLLMYLRYGNFLSSVVPDEQKSIGLSSSLDLRNSETVSTGYTS------- 199
Query 56 VNLFPLLAYQKIYQDFFRWSQWERANPSSYNVDYYSG--VSSSLVTVLPDYTSDYWKSDT 113
V++ PL YQKIY DFFR++QWE+ P +YN D+YSG V +SL T Y+ D
Sbjct 200 VHILPLATYQKIYADFFRFTQWEKNQPYTYNFDWYSGGNVLASLTT--QALAKKYYSDDN 257
Query 114 MFDLKYCNWNKDMLMGILPDSQFGECCYKSIFETPG--GDLKAG-FRTTDGKFISAVTNA 170
+F L+Y NW KDM MG++PDSQ G+ SI +T G G G + DG + +
Sbjct 258 LFTLRYANWPKDMFMGVMPDSQLGDV---SIVDTSGSEGTFPVGLYNFADGGSRAGLVAV 314
Query 171 PLTTENSSSGLSTPGVTSGSTVALKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSG 230
T+ + S L ++ S ++ L S FS+L LR AEA+Q+++E+SQ
Sbjct 315 SGTSPAAGSSLDMQTTSALSASTKYGVYAQQVAGLGSSFSILQLRMAEAVQKYREVSQFA 374
Query 231 DSDYREQIRKHFGVNLPQSLSNLCTYIGGISRNLDISEVVNNNLAAEGDTAVIAGKGVGA 290
D D R QI HFGV+L LS+ C Y+GG S N+D+SEVVN N+ + + A IAGKGVG
Sbjct 375 DQDARGQIMAHFGVSLSPVLSDKCMYLGGSSSNIDLSEVVNTNITGD-NIAEIAGKGVGT 433
Query 291 GNGSFTYTTDEHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGMEVLPMT 350
G GSF+ D + ++M IYH VPLLDY ITGQ LL T+ LP PE+D+IGM+ +
Sbjct 434 GQGSFSGNFDTYGIIMGIYHNVPLLDYVITGQPQNLLYTNTADLPFPEYDSIGMQTIQFG 493
Query 351 QIFNSPKASIVNLFN-----AGYNPRYFNWKTKLDVVNGAFTTTLKSWVSPVTESLLSGW 405
+ NS + + GY PR+F+ KT+ D V GAF +TLK+WV+P+ + L W
Sbjct 494 RFVNSKAVGWTSGVDYRTQTMGYLPRFFDVKTRYDEVLGAFRSTLKNWVAPLDPANLPQW 553
Query 406 FGFGYSQDDVNKDTKVV--LNYKFFKVNPSVLDPIFGVNADSTWDTDQLLVNSYIGCYVA 463
Q V K+ LNY FFKVNP VLD IF V DST DTDQ L Y+
Sbjct 554 L-----QTSVTSSGKLFLNLNYGFFKVNPRVLDSIFNVKCDSTIDTDQFLTTLYMDIKAV 608
Query 464 RNLSRDGVPY 473
RN DG+PY
Sbjct 609 RNFDYDGMPY 618
> Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1
Length=618
Score = 340 bits (873), Expect = 2e-111, Method: Compositional matrix adjust.
Identities = 213/512 (42%), Positives = 289/512 (56%), Gaps = 71/512 (14%)
Query 1 LGFSGADLAYKLLSYLGYGNFIPSPPSNSTRWWSTSLKKEASPTGYTQQYIQNNYVNLFP 60
G S ++ KL+SYLGYG E SP ++Y+ N + FP
Sbjct 139 FGVSRSEGFKKLVSYLGYG--------------------ETSP----EKYVDNLRCSAFP 174
Query 61 LLAYQKIYQDFFRWSQWERANPSSYNVDYYSGVSSSLVTVLPDYTSDYWKSDTMFDLKYC 120
L AYQKIYQD++R SQWE++ P +YN D+++G S+ +T + S +D++F+L+Y
Sbjct 175 LYAYQKIYQDYYRHSQWEKSKPWTYNCDFWNGEDSTPITATVELFSQS-PNDSVFELRYA 233
Query 121 NWNKDMLMGILPDSQFGECCYKSI-FETP------------GGDLKAGFRTTDGKFISA- 166
NWNKD+ MG LP+SQFG+ S+ F+ G++ G+ DG I +
Sbjct 234 NWNKDLWMGSLPNSQFGDVAGISLGFDASTMKVGVTGTALVKGNMPVGYGGKDGMGIRSQ 293
Query 167 --------VTNAPLTT---------ENS---SSGLSTPGVTSGSTVALKSPLISDLSA-L 205
+ +A T EN ++G G S + S L + LS L
Sbjct 294 SRLYNPVGINDAQQVTTVQQDVNNKENGYLFATGTDAFGRISNAAKINGSELFAQLSGQL 353
Query 206 QSQFSVLALRQAEALQRWKEISQSGDSDYREQIRKHFGVNLPQSLSNLCTYIGGISRNLD 265
+QFSVL LR AEALQ+WKEI+Q+ +Y Q++ HFGV+ S+ T I G ++D
Sbjct 354 DAQFSVLQLRAAEALQKWKEIAQANGQNYAAQVKAHFGVSTNPMQSHRSTRICGFDGSID 413
Query 266 ISEVVNNNLAAEGDTAVIAGKGVGAG--NGSFTYTTDEHCVVMCIYHAVPLLDYTITGQD 323
IS V N NL A D A+I GKG+G N +T +EH V+MCIYHA PLLDY TG D
Sbjct 414 ISAVENTNLTA--DEAIIRGKGLGGQRINDPSDFTCNEHGVIMCIYHATPLLDYVPTGPD 471
Query 324 GQLLVT-DAESLPIPEFDNIGMEVLPMTQIFNSPK-ASIVNLFNAGYNPRYFNWKTKLDV 381
QL+ T ES P+PEFD++GME LPM + NS IV AGY PRY +WKT +DV
Sbjct 472 LQLMSTVKGESWPVPEFDSLGMESLPMLSLVNSKAIGDIVARSYAGYVPRYISWKTSIDV 531
Query 382 VNGAFTTTLKSWVSPVTESLLSGWFGFGYSQDDVNKDTKVVLNYKFFKVNPSVLDPIFGV 441
V GAFT TLKSW +PV + +FG + + ++ +L+Y +FKVNPSVL+PIF V
Sbjct 532 VRGAFTDTLKSWTAPVDSDYMHVFFG-----EVIPQEGSPILSYTWFKVNPSVLNPIFAV 586
Query 442 NADSTWDTDQLLVNSYIGCYVARNLSRDGVPY 473
+ D +W+TDQLL N VARNLS DG+PY
Sbjct 587 SVDGSWNTDQLLCNCQFDVKVARNLSYDGMPY 618
> Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1
Length=602
Score = 338 bits (866), Expect = 1e-110, Method: Compositional matrix adjust.
Identities = 207/488 (42%), Positives = 276/488 (57%), Gaps = 42/488 (9%)
Query 2 GFSGADLAYKLLSYLGYGN-FIPSPPSNSTRWWSTSLKKEASPTGYTQQYIQNNYVNLFP 60
GFS A + KLL YL YGN + S PS + + L +S + +N N+ P
Sbjct 141 GFSKAATSAKLLRYLRYGNCYYTSDPSKVGKNKNFGL---SSKDDFNLLAAKNVSFNVLP 197
Query 61 LLAYQKIYQDFFRWSQWERANPSSYNVDYYSGVSS-SLVTVLPDYTSDYWKSDTMFDLKY 119
L AYQKIY D+FR+ QWE A P +YN DYY+G + + VT P+ ++W +D + L+Y
Sbjct 198 LAAYQKIYCDWFRFEQWENACPYTYNFDYYNGGNVFAGVTANPE---NFWSNDNILSLRY 254
Query 120 CNWNKDMLMGILPDSQFGECCYKSIFETPGGDLKAGFRTTDGKFISAVTNAPLTTENSSS 179
N+NKD+ MG++P SQFG ++ +L + R + +N N+SS
Sbjct 255 ANYNKDLFMGVMPSSQFGSVATVNVANFSSSNLSSPLRNLSNVGMVTASN------NASS 308
Query 180 GLSTPGVTSGSTVA------LKSPLISDLSALQSQFSVLALRQAEALQRWKEISQSGDSD 233
G +S T A L S + S LSA F +L+ R AEA Q+WKE++Q
Sbjct 309 GSPLILRSSQDTSAGDGFGILTSNIFSTLSA---SFDILSFRIAEATQKWKEVTQCAKQG 365
Query 234 YREQIRKHFGVNLPQSLSNLCTYIGGISRNLDISEVVNNNLAAEGDTAVIAGKGVGAGNG 293
Y+EQ+ HF V L ++LS+ C YIGG S + ISEV+N NL E A I GKGVG G
Sbjct 366 YKEQLEAHFNVKLSEALSDHCRYIGGTSSGVTISEVLNTNL--ESAAADIKGKGVGGSFG 423
Query 294 SFTYTTDEHCVVMCIYHAVPLLDYTITGQDGQLLVTDAESLPIPEFDNIGMEVLPMTQIF 353
S T+ T+EH ++MCIYHA P+LDY +GQD QLL T A +PIPEFD+IGME LP+ +F
Sbjct 424 SETFETNEHGILMCIYHATPVLDYLRSGQDLQLLSTLATDIPIPEFDHIGMEALPIETLF 483
Query 354 NSPKASIVNLFNA----GYNPRYFNWKTKLDVVNGAFTTTLKSWVSPVT--ESLLSGWFG 407
N L N+ GY+PRY +KT +D V+GAF TTL SWV+P+T E + F
Sbjct 484 NEQSTEATALINSIPVLGYSPRYIAYKTSVDWVSGAFETTLDSWVAPLTVNEQITKLLF- 542
Query 408 FGYSQDDVNKDTKVV--LNYKFFKVNPSVLDPIFGVNADSTWDTDQLLVNSYIGCYVARN 465
N D+ V +NY FFKV P VLDPIF TWD+DQ LVN +N
Sbjct 543 --------NPDSGSVYSMNYGFFKVTPRVLDPIFVQECTDTWDSDQFLVNVSFNVKPVQN 594
Query 466 LSRDGVPY 473
L +G+PY
Sbjct 595 LDYNGMPY 602
> Alpavirinae_Human_gut_33_005_Microviridae_AG0184_putative.VP1
Length=618
Score = 300 bits (767), Expect = 9e-96, Method: Compositional matrix adjust.
Identities = 197/513 (38%), Positives = 270/513 (53%), Gaps = 73/513 (14%)
Query 1 LGFSGADLAYKLLSYLGYGNFIPSPPSNSTRWWSTSLKKEASPTGYTQQYIQNNYVNLFP 60
GF+ + YKL + LG G PS + + N ++ FP
Sbjct 139 FGFARSSGFYKLFNSLGVGETDPS------------------------KTLANLRISAFP 174
Query 61 LLAYQKIYQDFFRWSQWERANPSSYNVDYYSGVSSSLVTVLPDYTSDYWKSDTMFDLKYC 120
AYQKIY D +R SQWE P +YN D+++G S+ V D D +D++F+L+Y
Sbjct 175 FYAYQKIYSDHYRNSQWEVNKPWTYNCDFWNGEDSTPVAFTKDLF-DTNPNDSVFELRYA 233
Query 121 NWNKDMLMGILPDSQFGECCYKSI-FETP--------------------GGDLKAGFRT- 158
NWNKD+ MG +P++QFG+ S+ F+T GG G R+
Sbjct 234 NWNKDLYMGAMPNTQFGDVAAVSLGFDTSTMKIGITGTAPVTGNMPVGYGGKDGMGLRSQ 293
Query 159 ---------TDGKFISAVTNAPLTTENS---SSGLSTPGVTSGSTVALKSPLISDLSA-L 205
D + ++ V EN ++G + G + S L + LS L
Sbjct 294 SRLYNPVGINDAQQVTTVQEEVNNKENGYLFATGTNAFGRIFNAAKISGSDLNAQLSGQL 353
Query 206 QSQFSVLALRQAEALQRWKEISQSGDSDYREQIRKHFGVNLPQSLSNLCTYIGGISRNLD 265
++FSVL LR AE LQ+WKEI+Q+ +Y Q++ HFGV+ S+ T + G ++D
Sbjct 354 DAKFSVLQLRAAECLQKWKEIAQANGQNYAAQVKAHFGVSPNPITSHRSTRVCGFDGSID 413
Query 266 ISEVVNNNLAAEGDTAVIAGKGVGA--GNGSFTYTTDEHCVVMCIYHAVPLLDYTITGQD 323
IS V N NL++ D A+I GKG+G N T+ T EH V+MCIYHAVPLLDY TG D
Sbjct 414 ISAVENTNLSS--DEAIIRGKGIGGYRVNKPETFETTEHGVLMCIYHAVPLLDYAPTGPD 471
Query 324 GQLLVT-DAESLPIPEFDNIGMEVLPMTQIFNSPKASIVNLFNA-GYNPRYFNWKTKLDV 381
Q + T D +S P+PE D+IG E LP + N+ + GY PRY +WKT +DV
Sbjct 472 LQFMTTVDGDSWPVPEMDSIGFEELPSYSLLNTNAVQPIKEPRPFGYVPRYISWKTSVDV 531
Query 382 VNGAFTTTLKSWVSPVTESLLSGWFGFGYSQDDVNKDTKVVLN-YKFFKVNPSVLDPIFG 440
V GAF TLKSW +P+ + L +F D+ N Y +FKVNPSV++PIFG
Sbjct 532 VRGAFIDTLKSWTAPIGQDYLKIYF------DNNNVPGGAHFGFYTWFKVNPSVVNPIFG 585
Query 441 VNADSTWDTDQLLVNSYIGCYVARNLSRDGVPY 473
V AD +W+TDQLLVN VARNLS DG+PY
Sbjct 586 VVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 618
Lambda K H a alpha
0.317 0.134 0.410 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 42522441