bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-44_CDS_annotation_glimmer3.pl_2_1
Length=598
Score E
Sequences producing significant alignments: (Bits) Value
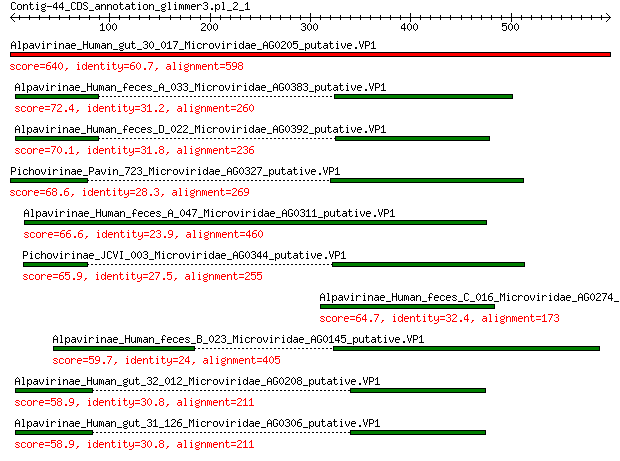
Alpavirinae_Human_gut_30_017_Microviridae_AG0205_putative.VP1 640 0.0
Alpavirinae_Human_feces_A_033_Microviridae_AG0383_putative.VP1 72.4 4e-15
Alpavirinae_Human_feces_D_022_Microviridae_AG0392_putative.VP1 70.1 2e-14
Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1 68.6 5e-14
Alpavirinae_Human_feces_A_047_Microviridae_AG0311_putative.VP1 66.6 2e-13
Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1 65.9 3e-13
Alpavirinae_Human_feces_C_016_Microviridae_AG0274_putative.VP1 64.7 1e-12
Alpavirinae_Human_feces_B_023_Microviridae_AG0145_putative.VP1 59.7 4e-11
Alpavirinae_Human_gut_32_012_Microviridae_AG0208_putative.VP1 58.9 6e-11
Alpavirinae_Human_gut_31_126_Microviridae_AG0306_putative.VP1 58.9 6e-11
> Alpavirinae_Human_gut_30_017_Microviridae_AG0205_putative.VP1
Length=647
Score = 640 bits (1651), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 363/663 (55%), Positives = 438/663 (66%), Gaps = 81/663 (12%)
Query 1 MGKQPFISHAVNGYSRYDVPESKAFTCTPGILYPVRIDFINARDRVSIEQGIDVRSNPLA 60
MGKQPFISH+VNGYSRYD+PE+KAF+ TPGI+YPVRI F+NARDRV++ QG+DVRSNPL
Sbjct 1 MGKQPFISHSVNGYSRYDMPENKAFSITPGIIYPVRIQFVNARDRVTLHQGVDVRSNPLG 60
Query 61 VPTFNPYTIRLHRFWVPLQLYHPELRTNSSKFDMNELSLNWICSTFPSAGSLN------- 113
VP+FNPY +RLHRFWVPLQLYHPE+R NSSKFDMN L+ N+I + G N
Sbjct 61 VPSFNPYVLRLHRFWVPLQLYHPEMRVNSSKFDMNNLTYNFIPGVVDNKGGGNYTSYMYP 120
Query 114 ----ADFFG--------ASYTNSLFSWLRVGNK--YNVGGTPLASVALPSSATLSQWS-- 157
A FF A+ NSL SWLR+ N +N G PL PS L + S
Sbjct 121 RSGTAAFFSQVMPFNCRAALPNSLMSWLRIANSPIFNYSGNPL-----PSGGVLFKTSPK 175
Query 158 ----NADTYLAYWDIVRNYYGYSQWGLYSFAWPMANKLVF----SSSVYTLDPDNSGDSR 209
NADTYL YWDIVRNYY YS WG++SFA P + VF SSSV ++ +
Sbjct 176 FLSVNADTYLGYWDIVRNYYSYSSWGVFSFAHPGTYRPVFYTGTSSSVTNVE--YRSQAS 233
Query 210 FFTQCYGNLEFLDAFYESQFYPS----SLNSSNNTFNRGNLFFQIVRSDLKSGGATS--- 262
+F Q YGNLEFLD ++E+ FYP + + ++NR +LF +I+RSDL + T+
Sbjct 234 YFWQRYGNLEFLDHYFETMFYPRDRIVAADRDELSWNRSDLFVEILRSDLFNKDTTAASP 293
Query 263 -----------------DGYPVVSTYPSTNLLGTTGLISQTLPstsssassGLAYFLVAH 305
YP P + TG +PS A++ FL AH
Sbjct 294 DFTKMPQMYPEAMNFNVPAYPYDVQEPKVDWNNGTG-TDVNVPSKVFFAATLNVPFLAAH 352
Query 306 PMAVVPSNPDRFSRLVPVG-SSSAVSMTGVTTIPQLAIASRLQEYKDLLGAGGSRYSDWL 364
PMAV PS+PDRFSRL+P G S+S V TG+ TIPQLA+A+RLQEYKDL+GA GSRYSDWL
Sbjct 353 PMAVCPSSPDRFSRLMPPGDSNSDVDFTGLKTIPQLAVATRLQEYKDLIGASGSRYSDWL 412
Query 365 ETFFASKIEHVDRPKLLFSASQTVNVQIVMNQAGDNNFSGNQP--LGQQGGSIAFNERLG 422
TFFASKIEHVDRPKLLFS+S VN Q+VMNQAG + F+G + LGQ GGSIAFN LG
Sbjct 413 YTFFASKIEHVDRPKLLFSSSVMVNSQVVMNQAGQSGFAGGEAAALGQMGGSIAFNTVLG 472
Query 423 RRQSYYFREPGYLIDMLSIRPVYYWSFIKPDYLNYSGSDYFNPIYNDIGYQDVPAFRIAF 482
R Q+YYF+EPGY+ DML+IRPVY+W+ I+PDYL Y G DYFNPIYNDIGYQDVP +R+ F
Sbjct 473 REQTYYFKEPGYIFDMLTIRPVYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPLWRLGF 532
Query 483 NGNPGA----SSASEPCFNEFRSSYDEVLGQLQAYYRSPAEGGTGAPLYSYWVQQR---A 535
A S A EPC+NEFRSSYDEVLG LQ+ A PL SYWVQQR
Sbjct 533 GWKSDAVSSVSVAKEPCYNEFRSSYDEVLGSLQSTLTPKAS----VPLQSYWVQQRDFYL 588
Query 536 VLTSSSTGSLPESYYYPVLFTDLSQVNSPFSSTVEDNFFVNLSYAVQKKNLVNKTFATRL 595
+ SS + S +LFT+L+ VN+PFSS +EDNFFVN+SY V KNLVNK+FATRL
Sbjct 589 IGLSSDLNEISPS----MLFTNLATVNNPFSSDMEDNFFVNMSYKVVVKNLVNKSFATRL 644
Query 596 SNR 598
S+R
Sbjct 645 SSR 647
> Alpavirinae_Human_feces_A_033_Microviridae_AG0383_putative.VP1
Length=590
Score = 72.4 bits (176), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 61/180 (34%), Positives = 89/180 (49%), Gaps = 15/180 (8%)
Query 324 GSSSAVSMTGVTTIPQLAIASRLQEYKDLLGAGGSRYSDWLETFFASKIEHVDR-PKLLF 382
S S+VSM +T ASR+Q Y DL AGG R SD+ E+ F K++ + P L
Sbjct 285 ASGSSVSMRNIT------FASRMQRYMDLAFAGGGRNSDFYESQFDVKLDQDNTCPAFLG 338
Query 383 SASQTVNVQIVMNQAGDNNFSGN-QPLGQQGGSIAFNERLGRRQSYYFREPGYLIDMLSI 441
S S +NV + G F N PLG G ++ R RR++Y+F + GY +++ SI
Sbjct 339 SDSFDMNVNTLYQTTG---FEDNSSPLGAFSGQLSGGTRF-RRRNYHFNDDGYFMEITSI 394
Query 442 RP-VYYWSFIKPDYLNYSGSDYFNPIYNDIGYQDVPAFRIAFNGNPGASSASEPCFNEFR 500
P VYY S+I P S + P ++I Q + A + G + A+ P + E R
Sbjct 395 VPRVYYPSYINPTSRQISLGQQYAPALDNIAMQGLKASTVF--GEVQSLGATNPSYAENR 452
Score = 39.3 bits (90), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 20/83 (24%), Positives = 40/83 (48%), Gaps = 0/83 (0%)
Query 6 FISHAVNGYSRYDVPESKAFTCTPGILYPVRIDFINARDRVSIEQGIDVRSNPLAVPTFN 65
F+S N SR+ + + + G L P + + A D S + G+ V++ P+ P
Sbjct 2 FLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFMG 61
Query 66 PYTIRLHRFWVPLQLYHPELRTN 88
++ F++P ++Y+ E + N
Sbjct 62 SVCVKKEYFFIPDRIYNIERQLN 84
> Alpavirinae_Human_feces_D_022_Microviridae_AG0392_putative.VP1
Length=589
Score = 70.1 bits (170), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 56/156 (36%), Positives = 78/156 (50%), Gaps = 13/156 (8%)
Query 325 SSSAVSMTGVTTIPQLAIASRLQEYKDLLGAGGSRYSDWLETFFASKIEHVDR-PKLLFS 383
S S+VSM +T ASR+Q Y DL AGG R SD+ E F K+ + P L S
Sbjct 285 SGSSVSMRNIT------FASRMQRYMDLAFAGGGRNSDFYEAQFDVKLNQDNTCPAFLGS 338
Query 384 ASQTVNVQIVMNQAGDNNFSGN-QPLGQQGGSIAFNERLGRRQSYYFREPGYLIDMLSIR 442
S +NV + G F N PLG G ++ R RR++Y+F + GY +++ SI
Sbjct 339 DSFDMNVNTLYQTTG---FEDNSSPLGAFSGQLSGGTRF-RRRNYHFNDDGYFMEITSIV 394
Query 443 P-VYYWSFIKPDYLNYSGSDYFNPIYNDIGYQDVPA 477
P VYY S+I P S + P ++I Q + A
Sbjct 395 PRVYYPSYINPTSRQISLGQQYAPALDNIAMQGLKA 430
Score = 37.4 bits (85), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 19/83 (23%), Positives = 40/83 (48%), Gaps = 0/83 (0%)
Query 6 FISHAVNGYSRYDVPESKAFTCTPGILYPVRIDFINARDRVSIEQGIDVRSNPLAVPTFN 65
F+S N SR+ + + + G L P + + A D S + G+ V++ P+ P
Sbjct 2 FLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVIAGDDFSFQPGVGVQALPIVAPFMG 61
Query 66 PYTIRLHRFWVPLQLYHPELRTN 88
++ F++P ++Y+ + + N
Sbjct 62 NVCVKKEYFFIPDRIYNIDRQLN 84
> Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1
Length=508
Score = 68.6 bits (166), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 59/210 (28%), Positives = 93/210 (44%), Gaps = 22/210 (10%)
Query 320 LVPVGSSSAVSMTGV--------------TTIPQLAIASRLQEYKDLLGAGGSRYSDWLE 365
L+ +G S+AV+ G TTI L A +LQE+ + GG+RY + +
Sbjct 238 LIDIGGSNAVNADGTPTMIAKTSTIDIEPTTINDLRRAFKLQEWLEKNARGGTRYIENIL 297
Query 366 TFFA--SKIEHVDRPKLLFSASQTVNVQIVMNQAGDNNFSGNQPLGQQGGSIAFNERLGR 423
T F S + + RP+ + V V V+N G + G P G G + G+
Sbjct 298 THFGVRSSDKRLQRPEYITGVKSPVVVSEVLNTTGQD---GGLPQGNMAGH-GISVTSGK 353
Query 424 RQSYYFREPGYLIDMLSIRP-VYYWSFIKPDYLNYSGSDYFNPIYNDIGYQDVPAFRI-A 481
SYY E GY+I ++S+ P Y I +L DYF P + +IG Q+V + A
Sbjct 354 SGSYYCEEHGYIIGIMSVMPKTAYQQGIPRTFLKTDSLDYFWPTFANIGEQEVAKQELYA 413
Query 482 FNGNPGASSASEPCFNEFRSSYDEVLGQLQ 511
+ N + P + E++ V G+ +
Sbjct 414 YTANANDTFGYVPRYAEYKYMPSRVAGEFR 443
Score = 24.6 bits (52), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 17/79 (22%), Positives = 32/79 (41%), Gaps = 2/79 (3%)
Query 1 MGKQPFISHAVNGYSR--YDVPESKAFTCTPGILYPVRIDFINARDRVSIEQGIDVRSNP 58
MG+ F S +N + +D+ + G L P+ D+ + +R P
Sbjct 1 MGQNLFNSIQLNKPKKNVFDLTHDVKLSTNMGQLTPILTLECVPGDKFDLSCESLIRFAP 60
Query 59 LAVPTFNPYTIRLHRFWVP 77
+ P + + +H F+VP
Sbjct 61 MIAPVMHRMDVTMHYFFVP 79
> Alpavirinae_Human_feces_A_047_Microviridae_AG0311_putative.VP1
Length=637
Score = 66.6 bits (161), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 110/519 (21%), Positives = 189/519 (36%), Gaps = 88/519 (17%)
Query 15 SRYDVPESKAFTCTPGILYPVRIDFINARDRVSIEQGIDVRSNPLAVPTFNPYTIRLHRF 74
S +D+ T PG L P + D + I+ +V ++P P F + + H +
Sbjct 26 STHDLSTVVRNTQAPGTLVPNLTLLVQKGDIIDIDIEANVLTHPTVGPLFGSFKLEHHIY 85
Query 75 WVPLQLYHPELRTNSSK--FDMNELSLNWICSTFPSAGSLNADFFGASYTNS-LFSWLRV 131
+P +LY+ L N +K DM+++ + + + + D S L ++L +
Sbjct 86 TIPFRLYNSWLHNNRTKIGLDMSQVKIPQLNVELKNTDNPTTDNEWTQVNPSCLLAYLGI 145
Query 132 GNKYNVGGTPLASVALPSSATLSQWSNADTYLAYWDIVRNYYGYSQWGLY---------- 181
++ GT + NA + Y+DI +NYY +Q +
Sbjct 146 RGYGSLIGTSAKYIG----------KNAVPLIGYYDIFKNYYANTQEEKFYTIGGVLPLN 195
Query 182 -------------SFAWPMANKLVFSSSVYTLD---------------PDNSGDSRFFTQ 213
+ A + +VFS+ D P + G T
Sbjct 196 VTYEGTEIPNSGNNIALSNGDTMVFSNVTNINDVVLIGSENLRYREWKPSDLGTVTGLTL 255
Query 214 -----CYGNLEFLDAFYE------SQFYPSSLNS--SNNTFNRGNLFFQIVRSDLKSGGA 260
GN L Y+ +Q+ SL++ N +GN+ F +
Sbjct 256 NIDKLLAGNEYILHKMYKKGMVDLNQWELESLDTLRDNILTTKGNVAFNV---------T 306
Query 261 TSDGYPVVSTYPSTNLLGTTGLISQTLPstsssassGLAYFLVAHPMAVVPSNPDRFSRL 320
D PV+ + TG + T P GLA + N D +
Sbjct 307 GKDSVPVLKQFNERFGEKQTGYLKTTAPQF------GLALKTYNSDLYQNWINTDWIEGV 360
Query 321 VPVGSSSAVSMT-GVTTIPQLAIASRLQEYKDLLGAGGSRYSDWLETFFASK--IEHVDR 377
+ S+V ++ G ++ L +A ++ + + G Y DWLET F +E +
Sbjct 361 NGINEISSVDVSDGSLSMDALNLAQKVYNMLNRIAVSGGTYRDWLETVFTGGEYMERCET 420
Query 378 PKLLFSASQTVNVQIVMNQAGDNNFSGNQPLGQQGG-SIAFNERLGRRQSYYFREPGYLI 436
P SQ +I+ + N+ + +PLG G I N++ G EPGY++
Sbjct 421 PIFEGGTSQ----EIIFQEVISNSATEQEPLGTLAGRGITTNKQRGGHVKIKVTEPGYIM 476
Query 437 DMLSIRP-VYYWSFIKPDYLNYSGSDYFNPIYNDIGYQD 474
+ SI P + Y + D + D P + IGYQD
Sbjct 477 CICSITPRIDYSQGNQWDTELKTMDDLHKPALDGIGYQD 515
> Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1
Length=505
Score = 65.9 bits (159), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 56/195 (29%), Positives = 88/195 (45%), Gaps = 12/195 (6%)
Query 322 PVGSSSAVSMTGVTTIPQLAIASRLQEYKDLLGAGGSRYSDWLETFF--ASKIEHVDRPK 379
P GS S TTI L A +LQE+ +LL GGSRY + ++TFF S + + RP+
Sbjct 255 PDGSLEVTS----TTITDLRRAFKLQEWLELLARGGSRYIEQIKTFFDVKSSDQRLQRPE 310
Query 380 LLFSASQTVNVQIVMNQAGDNNFSGNQPLGQQGGSIAFNERLGRRQSYYFREPGYLIDML 439
+ Q V + ++N G+ + P G G G YY E G++I +L
Sbjct 311 YITGTKQPVIISEILNTTGE---TAGLPQGNMSGH-GVAVGTGNVGKYYCEEHGFIIGIL 366
Query 440 SIRP-VYYWSFIKPDYLNYSGSDYFNPIYNDIGYQDVPAFRI-AFNGNPGASSASEPCFN 497
S+ P Y I YL D++ P + IG Q + I A++ + P +
Sbjct 367 SVTPNTAYQQGIPKHYLRTDKYDFYWPQFAHIGEQPIVNNEIYAYDPTGDETFGYTPRYA 426
Query 498 EFRSSYDEVLGQLQA 512
E++ V G+ ++
Sbjct 427 EYKYMPSRVAGEFRS 441
Score = 26.2 bits (56), Expect = 0.92, Method: Compositional matrix adjust.
Identities = 14/64 (22%), Positives = 28/64 (44%), Gaps = 0/64 (0%)
Query 14 YSRYDVPESKAFTCTPGILYPVRIDFINARDRVSIEQGIDVRSNPLAVPTFNPYTIRLHR 73
Y+ +D+ + G L P+ D +++ V+ PL P + + + +H
Sbjct 17 YNTFDLTHDVKMSGRMGELLPIMNLECIPGDNITLGADSMVKFAPLLAPVMHRFNVTMHY 76
Query 74 FWVP 77
F+VP
Sbjct 77 FFVP 80
> Alpavirinae_Human_feces_C_016_Microviridae_AG0274_putative.VP1
Length=641
Score = 64.7 bits (156), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 56/193 (29%), Positives = 85/193 (44%), Gaps = 23/193 (12%)
Query 310 VPSNPDRFSRLVPVGSSSAVSM-----------TGVT-TIPQLAIASRLQEYKDLLGAGG 357
VP +PD F ++ GSS + TG + +P+L + +++Q + D L G
Sbjct 318 VPYSPDLFGNIIKQGSSPTAEIEVIKALDSNTDTGFSIAVPELRLKTKIQNWMDRLFISG 377
Query 358 SRYSDWLETFFASK--IEHVDRPKLLFSASQTVNVQIVMNQAGDNNFSGNQPLGQQGGSI 415
R D T + +K +V++P L ++N V A + + LGQ +
Sbjct 378 GRVGDVFRTLWGTKSSAPYVNKPDFLGVWQASINPSNVRAMANGSASGEDANLGQLAACV 437
Query 416 A----FNERLGRRQSYYFREPG--YLIDMLSIRPVYYWSFIKPDYLNYSGSDYFNPIYND 469
F++ G YY +EPG LI ML P Y + PD + S D FNP N
Sbjct 438 DRYCDFSDHSG--IDYYAKEPGTFMLITMLVPEPAYSQG-LHPDLASISFGDDFNPELNG 494
Query 470 IGYQDVPAFRIAF 482
IG+Q VP R +
Sbjct 495 IGFQLVPRHRFSM 507
> Alpavirinae_Human_feces_B_023_Microviridae_AG0145_putative.VP1
Length=668
Score = 59.7 bits (143), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 62/275 (23%), Positives = 118/275 (43%), Gaps = 25/275 (9%)
Query 323 VGSSSAVSMTGVT-TIPQLAIASRLQEYKDLLGAGGSRYSDWLETFFASKIE-HVDRPKL 380
+ + +A+S G TI QL ++ ++ + + + G Y DW+ET + S+ H + P
Sbjct 401 ISAITAISTEGNKFTIDQLNLSKKVYDMLNRIAISGGTYQDWVETVYTSEWNMHTETPVY 460
Query 381 LFSASQTVNVQIVMNQAGDNNFSGNQPLGQQGGSIAFNERLGRRQSYYFREPGYLIDMLS 440
S + Q V++ N+ + +PLG G + + G + EP Y++ + S
Sbjct 461 EGGMSAEIEFQEVVS----NSATEEEPLGTLAGRGFASNKKGGQLHIKVTEPCYIMGIAS 516
Query 441 IRP-VYY-----WSFIKPDYLNYSGSDYFNPIYNDIGYQDVPAFRIAFNGNPGASSASEP 494
I P V Y W D ++ D P + IGYQD+ ++ + + +P
Sbjct 517 ITPRVDYCQGNDWDITSLDTMD----DIHKPQLDSIGYQDLMQEQMNAQASRNLAVGKQP 572
Query 495 CFNEFRSSYDEVLGQLQAYYRSPAEGGTGAPLYSYWVQQRAVLTSSSTGSLPESYYYPVL 554
+ + +S+++ G + EG L Y+ + + + TG Y
Sbjct 573 SWINYMTSFNKTYG---TFANEDGEGEAFMVLNRYF-DIKEIDAGTETGV---KVYNTST 625
Query 555 FTDLSQVNSPFSSTVED--NFFVNLSYAVQKKNLV 587
+ D SQ N F+ T NF+V L + ++ + ++
Sbjct 626 YIDPSQYNYIFAETGTKSMNFWVQLGFGIEARRVM 660
Score = 34.7 bits (78), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 35/142 (25%), Positives = 59/142 (42%), Gaps = 12/142 (8%)
Query 44 DRVSIEQGIDVRSNPLAVPTFNPYTIRLHRFWVPLQLYHPELRTNSSK--FDMNELSLNW 101
D I+ V ++P P F Y +++ F VP +LY L N+ DM+++ +
Sbjct 57 DTFDIKIDNKVLTHPTVGPLFGSYKLQVDLFTVPFRLYIAMLHNNALNVGMDMSKVKIPI 116
Query 102 ICSTFPSAGSLNADFFGASYTNSLFSWLRVGNKYNVGGTPLASVALPSSATLSQWSNADT 161
+L+ D +S + +F++L G + +S +Q NA
Sbjct 117 YRVGQKQGTNLSKDLLYSS--SCIFTYL--------GNRGQETKKNSTSNVNTQDFNAIP 166
Query 162 YLAYWDIVRNYYGYSQWGLYSF 183
L Y DI +NYY Q + F
Sbjct 167 LLGYLDIFKNYYANKQEEKFKF 188
> Alpavirinae_Human_gut_32_012_Microviridae_AG0208_putative.VP1
Length=604
Score = 58.9 bits (141), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 43/136 (32%), Positives = 68/136 (50%), Gaps = 5/136 (4%)
Query 340 LAIASRLQEYKDLLGAGGSRYSDWLETFF-ASKIEHVDRPKLLFSASQTVNVQIVMNQAG 398
+A S +Q + DL AGGSRYSD++ + F S+++H P L S Q + ++ G
Sbjct 300 IAAQSHIQRWLDLALAGGSRYSDYINSQFDVSRLKHTTSPLYLGSDRQYLGSNVIYQTTG 359
Query 399 DNNFSGNQPLGQQGGSIAFNERLGRRQSYYFREPGYLIDMLSIRP-VYYWSFIKPDYLNY 457
+ + PLG G + E RR+SY+F E GY + M S+ P V Y + P
Sbjct 360 AGDSA--SPLGAFAGQSSGGETF-RRRSYHFGENGYFVVMASLVPDVVYSRGMDPFNREK 416
Query 458 SGSDYFNPIYNDIGYQ 473
+ D + P ++I +
Sbjct 417 TLGDVYVPALDNIAME 432
Score = 37.7 bits (86), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/77 (29%), Positives = 37/77 (48%), Gaps = 0/77 (0%)
Query 6 FISHAVNGYSRYDVPESKAFTCTPGILYPVRIDFINARDRVSIEQGIDVRSNPLAVPTFN 65
F++ + N S ++ T PG L P+ I A D + E V++ P+ P N
Sbjct 5 FLTRSRNKKSNVNLSHVSPTTIAPGNLVPISFTRIFAGDDLRFEPSAFVQAMPMNAPLVN 64
Query 66 PYTIRLHRFWVPLQLYH 82
+ + L F+VP +LY+
Sbjct 65 GFKLCLEYFFVPDRLYN 81
> Alpavirinae_Human_gut_31_126_Microviridae_AG0306_putative.VP1
Length=604
Score = 58.9 bits (141), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 43/136 (32%), Positives = 68/136 (50%), Gaps = 5/136 (4%)
Query 340 LAIASRLQEYKDLLGAGGSRYSDWLETFF-ASKIEHVDRPKLLFSASQTVNVQIVMNQAG 398
+A S +Q + DL AGGSRYSD++ + F S+++H P L S Q + ++ G
Sbjct 300 IAAQSHIQRWLDLALAGGSRYSDYINSQFDVSRLKHTTSPLYLGSDRQYLGSNVIYQTTG 359
Query 399 DNNFSGNQPLGQQGGSIAFNERLGRRQSYYFREPGYLIDMLSIRP-VYYWSFIKPDYLNY 457
+ + PLG G + E RR+SY+F E GY + M S+ P V Y + P
Sbjct 360 AGDSA--SPLGAFAGQSSGGETF-RRRSYHFGENGYFVVMASLVPDVVYSRGMDPFNREK 416
Query 458 SGSDYFNPIYNDIGYQ 473
+ D + P ++I +
Sbjct 417 TLGDVYVPALDNIAME 432
Score = 37.7 bits (86), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/77 (29%), Positives = 37/77 (48%), Gaps = 0/77 (0%)
Query 6 FISHAVNGYSRYDVPESKAFTCTPGILYPVRIDFINARDRVSIEQGIDVRSNPLAVPTFN 65
F++ + N S ++ T PG L P+ I A D + E V++ P+ P N
Sbjct 5 FLTRSRNKKSNVNLSHVSPTTIAPGNLVPISFTRIFAGDDLRFEPSAFVQAMPMNAPLVN 64
Query 66 PYTIRLHRFWVPLQLYH 82
+ + L F+VP +LY+
Sbjct 65 GFKLCLEYFFVPDRLYN 81
Lambda K H a alpha
0.318 0.134 0.410 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 54962884