bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-3_CDS_annotation_glimmer3.pl_2_7
Length=166
Score E
Sequences producing significant alignments: (Bits) Value
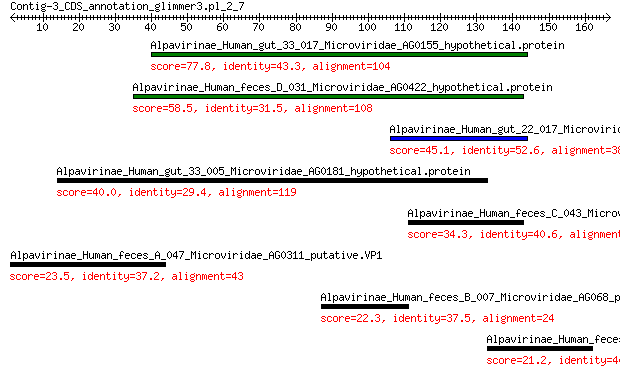
Alpavirinae_Human_gut_33_017_Microviridae_AG0155_hypothetical.p... 77.8 6e-20
Alpavirinae_Human_feces_D_031_Microviridae_AG0422_hypothetical.... 58.5 3e-13
Alpavirinae_Human_gut_22_017_Microviridae_AG0398_hypothetical.p... 45.1 6e-09
Alpavirinae_Human_gut_33_005_Microviridae_AG0181_hypothetical.p... 40.0 3e-06
Alpavirinae_Human_feces_C_043_Microviridae_AG0323_hypothetical.... 34.3 6e-05
Alpavirinae_Human_feces_A_047_Microviridae_AG0311_putative.VP1 23.5 1.1
Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1 22.3 2.9
Alpavirinae_Human_feces_A_032_Microviridae_AG0219_putative.VP2 21.2 5.7
> Alpavirinae_Human_gut_33_017_Microviridae_AG0155_hypothetical.protein
Length=171
Score = 77.8 bits (190), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 45/105 (43%), Positives = 65/105 (62%), Gaps = 5/105 (5%)
Query 40 DPVEQFRFETETFGESVSYRLRSDVSMLLHAADLAKRAGVSTVQRFLDSK-SPRSSSLQE 98
D EQ R E + S R SD+ ++LH DLA RAGV +F SK SP S +Q+
Sbjct 44 DSTEQLRVEIDDTDASRPVRYTSDIRLILHNKDLASRAGVDVASKFGQSKQSP--SQIQQ 101
Query 99 QLDKLNPSDDELLSMVKSRHLQHPSEILAWIDSINELAEDMRSEA 143
+D + SD++LL+ V+SR++Q PSEILAW ++ AE++ S+A
Sbjct 102 IMDTM--SDEDLLATVRSRYIQSPSEILAWSKELSAYAENLESQA 144
> Alpavirinae_Human_feces_D_031_Microviridae_AG0422_hypothetical.protein
Length=150
Score = 58.5 bits (140), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 34/108 (31%), Positives = 57/108 (53%), Gaps = 8/108 (7%)
Query 35 ISLEVDPVEQFRFETETFGESVSYRLRSDVSMLLHAADLAKRAGVSTVQRFLDSKSPRSS 94
I +V PVE R+ + G R SDV++L++A L + G + + P+ S
Sbjct 26 IYCQVGPVEMLRYVKDDDG---VIRYVSDVNLLMNAERLRNQIGEESYLNLIRGIQPKKS 82
Query 95 SLQEQLDKLNPSDDELLSMVKSRHLQHPSEILAWIDSINELAEDMRSE 142
+ +D++L + +KSR +Q PSE+LAWI+S+ + +RSE
Sbjct 83 PYDNKY-----TDEQLFTAIKSRFIQTPSEVLAWIESLGSAGDSIRSE 125
> Alpavirinae_Human_gut_22_017_Microviridae_AG0398_hypothetical.protein
Length=69
Score = 45.1 bits (105), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 20/38 (53%), Positives = 29/38 (76%), Gaps = 0/38 (0%)
Query 106 SDDELLSMVKSRHLQHPSEILAWIDSINELAEDMRSEA 143
SDD+LL+ V+SRH+Q PSEI+AW ++ AE + S+A
Sbjct 5 SDDDLLATVRSRHIQAPSEIIAWSKELSAYAEHLESQA 42
> Alpavirinae_Human_gut_33_005_Microviridae_AG0181_hypothetical.protein
Length=205
Score = 40.0 bits (92), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 35/128 (27%), Positives = 59/128 (46%), Gaps = 18/128 (14%)
Query 14 GFREFTPTREIS--------TFPSTRIGEISL-EVDPVEQFRFETETFGESVSYRLRSDV 64
GF +++ R+IS S + + L E +PV F F+ G S SD
Sbjct 49 GFAKYSLNRDISDHSQSCRCVMTSEMLAQGILPEPNPVGDFLFDHNADG---SVTFCSDY 105
Query 65 SMLLHAADLAKRAGVSTVQRFLDSKSPRSSSLQEQLDKLNPSDDELLSMVKSRHLQHPSE 124
+L + V ++R+++S PRSS+ N +DD LL K R++Q +E
Sbjct 106 GILFGQKAIDNMNQVQ-LRRYMNSLVPRSSNYTR-----NYNDDFLLDYCKDRNIQSATE 159
Query 125 ILAWIDSI 132
+ +W+D +
Sbjct 160 MASWLDHL 167
> Alpavirinae_Human_feces_C_043_Microviridae_AG0323_hypothetical.protein
Length=59
Score = 34.3 bits (77), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 13/32 (41%), Positives = 25/32 (78%), Gaps = 0/32 (0%)
Query 111 LSMVKSRHLQHPSEILAWIDSINELAEDMRSE 142
+ +KSR+LQ PSE+ AW++++ + A+ +RS+
Sbjct 1 METIKSRYLQSPSEVRAWLETLVDKADVVRSD 32
> Alpavirinae_Human_feces_A_047_Microviridae_AG0311_putative.VP1
Length=637
Score = 23.5 bits (49), Expect = 1.1, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 21/43 (49%), Gaps = 10/43 (23%)
Query 1 MIMKEKKELCFGCGFREFTPTREISTFPSTRIGEISLEVDPVE 43
M+M EL G TPT S RIG++S +DPV+
Sbjct 568 MVMNRNYELQINGGP---TPT-------SIRIGDLSTYIDPVK 600
> Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1
Length=780
Score = 22.3 bits (46), Expect = 2.9, Method: Composition-based stats.
Identities = 9/24 (38%), Positives = 14/24 (58%), Gaps = 0/24 (0%)
Query 87 DSKSPRSSSLQEQLDKLNPSDDEL 110
DS P+ + +E+LD NP D +
Sbjct 644 DSILPKWLTYRERLDSFNPEFDHI 667
> Alpavirinae_Human_feces_A_032_Microviridae_AG0219_putative.VP2
Length=355
Score = 21.2 bits (43), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 13/29 (45%), Positives = 14/29 (48%), Gaps = 0/29 (0%)
Query 133 NELAEDMRSEALKQTAENGSICPIVLVKK 161
N AE +RSE Q EN I L KK
Sbjct 148 NAQAEKIRSETTTQKVENMVGIGIDLAKK 176
Lambda K H a alpha
0.315 0.130 0.362 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 11626264