bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
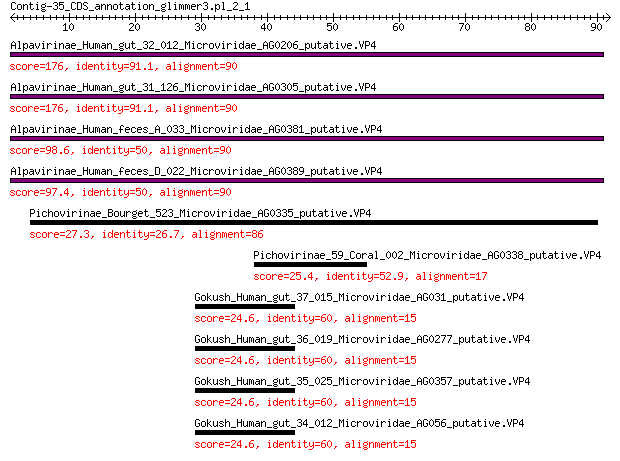
Query= Contig-35_CDS_annotation_glimmer3.pl_2_1
Length=91
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_gut_32_012_Microviridae_AG0206_putative.VP4 176 2e-57
Alpavirinae_Human_gut_31_126_Microviridae_AG0305_putative.VP4 176 2e-57
Alpavirinae_Human_feces_A_033_Microviridae_AG0381_putative.VP4 98.6 9e-28
Alpavirinae_Human_feces_D_022_Microviridae_AG0389_putative.VP4 97.4 2e-27
Pichovirinae_Bourget_523_Microviridae_AG0335_putative.VP4 27.3 0.015
Pichovirinae_59_Coral_002_Microviridae_AG0338_putative.VP4 25.4 0.075
Gokush_Human_gut_37_015_Microviridae_AG031_putative.VP4 24.6 0.15
Gokush_Human_gut_36_019_Microviridae_AG0277_putative.VP4 24.6 0.15
Gokush_Human_gut_35_025_Microviridae_AG0357_putative.VP4 24.6 0.15
Gokush_Human_gut_34_012_Microviridae_AG056_putative.VP4 24.6 0.15
> Alpavirinae_Human_gut_32_012_Microviridae_AG0206_putative.VP4
Length=306
Score = 176 bits (447), Expect = 2e-57, Method: Compositional matrix adjust.
Identities = 82/90 (91%), Positives = 86/90 (96%), Gaps = 0/90 (0%)
Query 1 VSHGFGRLSKEDIKIMRENMLRSSTSWFCVYINNFRYSIPRYWRTACFTKDEIKCRNDSL 60
VSHGFGRLSKEDIKIMRENMLRSS SWFCVY+NNFRYSIPRYWRTACFTKDEIKCRNDSL
Sbjct 216 VSHGFGRLSKEDIKIMRENMLRSSASWFCVYVNNFRYSIPRYWRTACFTKDEIKCRNDSL 275
Query 61 IPAILWSIVQQRYRGLSPLNQRLIYYSLLW 90
IPAILWSIVQQ+YRG SP+ Q+ IYYSLLW
Sbjct 276 IPAILWSIVQQKYRGYSPVQQQQIYYSLLW 305
> Alpavirinae_Human_gut_31_126_Microviridae_AG0305_putative.VP4
Length=306
Score = 176 bits (447), Expect = 2e-57, Method: Compositional matrix adjust.
Identities = 82/90 (91%), Positives = 86/90 (96%), Gaps = 0/90 (0%)
Query 1 VSHGFGRLSKEDIKIMRENMLRSSTSWFCVYINNFRYSIPRYWRTACFTKDEIKCRNDSL 60
VSHGFGRLSKEDIKIMRENMLRSS SWFCVY+NNFRYSIPRYWRTACFTKDEIKCRNDSL
Sbjct 216 VSHGFGRLSKEDIKIMRENMLRSSASWFCVYVNNFRYSIPRYWRTACFTKDEIKCRNDSL 275
Query 61 IPAILWSIVQQRYRGLSPLNQRLIYYSLLW 90
IPAILWSIVQQ+YRG SP+ Q+ IYYSLLW
Sbjct 276 IPAILWSIVQQKYRGYSPVQQQQIYYSLLW 305
> Alpavirinae_Human_feces_A_033_Microviridae_AG0381_putative.VP4
Length=307
Score = 98.6 bits (244), Expect = 9e-28, Method: Compositional matrix adjust.
Identities = 45/90 (50%), Positives = 61/90 (68%), Gaps = 0/90 (0%)
Query 1 VSHGFGRLSKEDIKIMRENMLRSSTSWFCVYINNFRYSIPRYWRTACFTKDEIKCRNDSL 60
VSHGFGRLS+ + +R M+ WF + I+N YSIPRY++ ACFTKD+I+CRNDS
Sbjct 217 VSHGFGRLSESERDALRAYMMTGCKQWFSILIDNHPYSIPRYYKLACFTKDQIRCRNDSF 276
Query 61 IPAILWSIVQQRYRGLSPLNQRLIYYSLLW 90
IP ++W V + Y S ++LI YS+LW
Sbjct 277 IPQLVWEYVLRTYPTYSYYKKQLIKYSILW 306
> Alpavirinae_Human_feces_D_022_Microviridae_AG0389_putative.VP4
Length=307
Score = 97.4 bits (241), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 45/90 (50%), Positives = 61/90 (68%), Gaps = 0/90 (0%)
Query 1 VSHGFGRLSKEDIKIMRENMLRSSTSWFCVYINNFRYSIPRYWRTACFTKDEIKCRNDSL 60
VSHGFGRLS+ + +R M+ WF + I+N YSIPRY++ ACFTKD+I+CRNDSL
Sbjct 217 VSHGFGRLSESEKDTLRAYMMTGCKQWFSILIDNHPYSIPRYYKLACFTKDQIRCRNDSL 276
Query 61 IPAILWSIVQQRYRGLSPLNQRLIYYSLLW 90
IP ++W V + Y S ++LI S+LW
Sbjct 277 IPQLVWEYVLRTYPTYSYYKKQLIKQSILW 306
> Pichovirinae_Bourget_523_Microviridae_AG0335_putative.VP4
Length=291
Score = 27.3 bits (59), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 23/86 (27%), Positives = 39/86 (45%), Gaps = 5/86 (6%)
Query 4 GFGRLSKEDIKIMRENMLRSSTSWFCVYINNFRYSIPRYWRTACFTKDEIKCRNDSLIPA 63
G LS++ I+ R ++ R+ F + + S+PRY+R +T+ E + + D L
Sbjct 188 GLNYLSEKIIRYHRSDIERN----FITLEDGKKISLPRYFREKIWTEQERRIQADKLAQK 243
Query 64 ILWSIVQQRYRGLSPLNQRLIYYSLL 89
+ QQ+ NQ L Y L
Sbjct 244 -FKELEQQKETEYYTKNQTLEGYEQL 268
> Pichovirinae_59_Coral_002_Microviridae_AG0338_putative.VP4
Length=274
Score = 25.4 bits (54), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 9/17 (53%), Positives = 13/17 (76%), Gaps = 0/17 (0%)
Query 38 SIPRYWRTACFTKDEIK 54
S+PRY++ FTK E+K
Sbjct 212 SMPRYYKEKIFTKQELK 228
> Gokush_Human_gut_37_015_Microviridae_AG031_putative.VP4
Length=315
Score = 24.6 bits (52), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 9/15 (60%), Positives = 11/15 (73%), Gaps = 0/15 (0%)
Query 29 CVYINNFRYSIPRYW 43
C INN R+ IPRY+
Sbjct 235 CCLINNKRFKIPRYY 249
> Gokush_Human_gut_36_019_Microviridae_AG0277_putative.VP4
Length=315
Score = 24.6 bits (52), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 9/15 (60%), Positives = 11/15 (73%), Gaps = 0/15 (0%)
Query 29 CVYINNFRYSIPRYW 43
C INN R+ IPRY+
Sbjct 235 CCLINNKRFKIPRYY 249
> Gokush_Human_gut_35_025_Microviridae_AG0357_putative.VP4
Length=315
Score = 24.6 bits (52), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 9/15 (60%), Positives = 11/15 (73%), Gaps = 0/15 (0%)
Query 29 CVYINNFRYSIPRYW 43
C INN R+ IPRY+
Sbjct 235 CCLINNKRFKIPRYY 249
> Gokush_Human_gut_34_012_Microviridae_AG056_putative.VP4
Length=315
Score = 24.6 bits (52), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 9/15 (60%), Positives = 11/15 (73%), Gaps = 0/15 (0%)
Query 29 CVYINNFRYSIPRYW 43
C INN R+ IPRY+
Sbjct 235 CCLINNKRFKIPRYY 249
Lambda K H a alpha
0.328 0.139 0.466 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4169376