bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
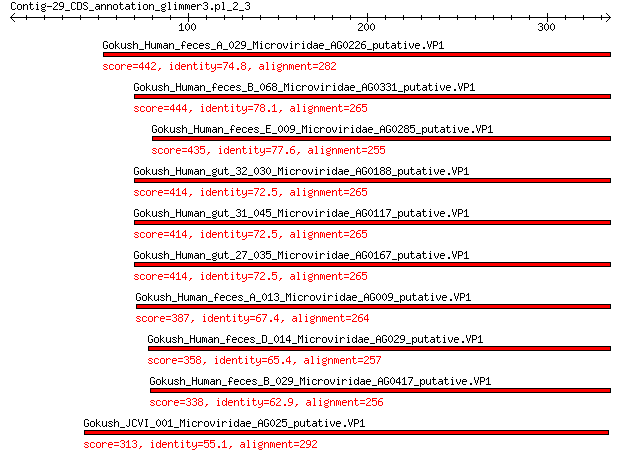
Query= Contig-29_CDS_annotation_glimmer3.pl_2_3
Length=334
Score E
Sequences producing significant alignments: (Bits) Value
Gokush_Human_feces_A_029_Microviridae_AG0226_putative.VP1 442 3e-155
Gokush_Human_feces_B_068_Microviridae_AG0331_putative.VP1 444 1e-154
Gokush_Human_feces_E_009_Microviridae_AG0285_putative.VP1 435 7e-151
Gokush_Human_gut_32_030_Microviridae_AG0188_putative.VP1 414 1e-142
Gokush_Human_gut_31_045_Microviridae_AG0117_putative.VP1 414 1e-142
Gokush_Human_gut_27_035_Microviridae_AG0167_putative.VP1 414 1e-142
Gokush_Human_feces_A_013_Microviridae_AG009_putative.VP1 387 4e-132
Gokush_Human_feces_D_014_Microviridae_AG029_putative.VP1 358 6e-121
Gokush_Human_feces_B_029_Microviridae_AG0417_putative.VP1 338 9e-114
Gokush_JCVI_001_Microviridae_AG025_putative.VP1 313 1e-103
> Gokush_Human_feces_A_029_Microviridae_AG0226_putative.VP1
Length=470
Score = 442 bits (1138), Expect = 3e-155, Method: Compositional matrix adjust.
Identities = 211/282 (75%), Positives = 235/282 (83%), Gaps = 3/282 (1%)
Query 53 GVGFDAPTSRDGSMYLDNLWAIQSGNVTAATINQLRMAFQIQKLYEKDARGGTRYIEILK 112
GVG R S L A+ G V+ ATINQLR+AFQIQKLYE+DARGGTRYIEILK
Sbjct 192 GVGTQGQLGRGTS---SGLIAVDDGGVSMATINQLRLAFQIQKLYERDARGGTRYIEILK 248
Query 113 SHFGVTSPDARLQRPEYLGGNRIPVNINQVVQSSATQSSGTPLGDTAAFSVTTDVHGDFI 172
SHFGVTSPDARLQRPEYLGGNRIP+ INQVVQ+S T S TP G T A+S+TTDVH +F
Sbjct 249 SHFGVTSPDARLQRPEYLGGNRIPITINQVVQNSGTMSGETPQGTTTAYSLTTDVHQEFT 308
Query 173 KSFVEHGFVIGIMVARYDHTYQQGLERFWSRRDRLDYYFPVFANIGEQPILNKEIYAQGT 232
KSFVEHGF+IG+MVARYDHTYQQGLERFWSR+DR D+Y+PVFANIGEQ ILNKEIYA G
Sbjct 309 KSFVEHGFIIGVMVARYDHTYQQGLERFWSRKDRFDFYWPVFANIGEQAILNKEIYATGK 368
Query 233 VQDNEVFGYQEAWADYRYKPSRVAGEMRSKAPTSLDVWHLADEYTQLPKLSDAWIREDKT 292
D+EVFGYQEAWADYRYKPSRV+GEMRS A T LD WHLAD+Y LP LSD+WIRE+
Sbjct 369 DSDSEVFGYQEAWADYRYKPSRVSGEMRSNAKTPLDSWHLADDYDVLPTLSDSWIREESN 428
Query 293 NVDRVLAVTSAVSNQMFADLYIQCKATRPMPMYSIPGLIDHH 334
NV+RVLAVTS VSNQ+F DLY+Q + TRPMP+YSIPGLIDHH
Sbjct 429 NVNRVLAVTSEVSNQLFCDLYVQNRTTRPMPVYSIPGLIDHH 470
> Gokush_Human_feces_B_068_Microviridae_AG0331_putative.VP1
Length=547
Score = 444 bits (1141), Expect = 1e-154, Method: Compositional matrix adjust.
Identities = 207/265 (78%), Positives = 232/265 (88%), Gaps = 0/265 (0%)
Query 70 NLWAIQSGNVTAATINQLRMAFQIQKLYEKDARGGTRYIEILKSHFGVTSPDARLQRPEY 129
NL A G AATINQLR+AFQIQKLYE+DARGGTRYIEILKSHFGVTSPDARLQRPEY
Sbjct 283 NLIAKFDGVSQAATINQLRLAFQIQKLYERDARGGTRYIEILKSHFGVTSPDARLQRPEY 342
Query 130 LGGNRIPVNINQVVQSSATQSSGTPLGDTAAFSVTTDVHGDFIKSFVEHGFVIGIMVARY 189
LGGNRIP+NINQVVQSS+T +SGTP G+TAA+S+T+D H DF KSFVEHGF+IG+MVARY
Sbjct 343 LGGNRIPININQVVQSSSTDASGTPQGNTAAYSLTSDNHSDFTKSFVEHGFLIGVMVARY 402
Query 190 DHTYQQGLERFWSRRDRLDYYFPVFANIGEQPILNKEIYAQGTVQDNEVFGYQEAWADYR 249
HTYQQGLERFWSR+DR DYYFPVFANIGEQ I NKEIYAQGTV+D+EVFGYQEAWADYR
Sbjct 403 RHTYQQGLERFWSRKDRFDYYFPVFANIGEQAIKNKEIYAQGTVKDDEVFGYQEAWADYR 462
Query 250 YKPSRVAGEMRSKAPTSLDVWHLADEYTQLPKLSDAWIREDKTNVDRVLAVTSAVSNQMF 309
Y+P+RV GEMRS AP SLDVWHL D+Y LP LSD+WIRED V+RVLAV+ VS Q+F
Sbjct 463 YRPNRVTGEMRSSAPQSLDVWHLGDDYESLPSLSDSWIREDSKTVNRVLAVSDNVSAQLF 522
Query 310 ADLYIQCKATRPMPMYSIPGLIDHH 334
D+Y++ TRPMP+YSIPGLIDHH
Sbjct 523 CDIYVRNLCTRPMPLYSIPGLIDHH 547
> Gokush_Human_feces_E_009_Microviridae_AG0285_putative.VP1
Length=559
Score = 435 bits (1118), Expect = 7e-151, Method: Compositional matrix adjust.
Identities = 198/255 (78%), Positives = 224/255 (88%), Gaps = 0/255 (0%)
Query 80 TAATINQLRMAFQIQKLYEKDARGGTRYIEILKSHFGVTSPDARLQRPEYLGGNRIPVNI 139
T+ATINQLR+AFQIQKLYEKDARGGTRY EILK+HFGVTSPD+RLQRPEYLGGNR+P+NI
Sbjct 305 TSATINQLRLAFQIQKLYEKDARGGTRYTEILKTHFGVTSPDSRLQRPEYLGGNRVPINI 364
Query 140 NQVVQSSATQSSGTPLGDTAAFSVTTDVHGDFIKSFVEHGFVIGIMVARYDHTYQQGLER 199
NQVVQ+SAT TPLG+ A +SVT+D H DF +SF EHGFVIG+MVARYDHTYQQG+ER
Sbjct 365 NQVVQNSATVEGETPLGNVAGYSVTSDTHSDFRQSFTEHGFVIGVMVARYDHTYQQGIER 424
Query 200 FWSRRDRLDYYFPVFANIGEQPILNKEIYAQGTVQDNEVFGYQEAWADYRYKPSRVAGEM 259
FWSR+ R DYY+PV ANIGEQ +LNKEIYAQGT +D+EVFGYQEAW DYRYKP+RV GEM
Sbjct 425 FWSRKTRFDYYWPVLANIGEQAVLNKEIYAQGTAEDDEVFGYQEAWGDYRYKPNRVTGEM 484
Query 260 RSKAPTSLDVWHLADEYTQLPKLSDAWIREDKTNVDRVLAVTSAVSNQMFADLYIQCKAT 319
RS+ SLDVWHL D+YT+LP LS WI EDKTNVDRVLAVTS +NQ+FADLYI + T
Sbjct 485 RSQYAQSLDVWHLGDDYTKLPSLSSEWIVEDKTNVDRVLAVTSTNANQLFADLYINNQTT 544
Query 320 RPMPMYSIPGLIDHH 334
RPMPMYSIPGL+DHH
Sbjct 545 RPMPMYSIPGLVDHH 559
> Gokush_Human_gut_32_030_Microviridae_AG0188_putative.VP1
Length=562
Score = 414 bits (1063), Expect = 1e-142, Method: Compositional matrix adjust.
Identities = 192/265 (72%), Positives = 222/265 (84%), Gaps = 0/265 (0%)
Query 70 NLWAIQSGNVTAATINQLRMAFQIQKLYEKDARGGTRYIEILKSHFGVTSPDARLQRPEY 129
NLWA+ GN AATINQLR+AFQIQK YE+ ARGG+RY E+++S FGVTSPDARLQRPEY
Sbjct 298 NLWAVADGNAAAATINQLRLAFQIQKFYERQARGGSRYTEVVRSFFGVTSPDARLQRPEY 357
Query 130 LGGNRIPVNINQVVQSSATQSSGTPLGDTAAFSVTTDVHGDFIKSFVEHGFVIGIMVARY 189
LGGNR+P+N+NQ+VQ S TQS TP G S+TTD H DF KSF EHG +IG+MVARY
Sbjct 358 LGGNRVPINVNQIVQQSGTQSGTTPQGTVVGQSLTTDKHSDFTKSFTEHGLIIGVMVARY 417
Query 190 DHTYQQGLERFWSRRDRLDYYFPVFANIGEQPILNKEIYAQGTVQDNEVFGYQEAWADYR 249
DHTYQQGL R WSR+D+ D+Y+PVFANIGEQ I NKEI+AQG +DNEVFGYQEAWA+YR
Sbjct 418 DHTYQQGLNRLWSRKDKFDFYWPVFANIGEQAIKNKEIFAQGNDKDNEVFGYQEAWAEYR 477
Query 250 YKPSRVAGEMRSKAPTSLDVWHLADEYTQLPKLSDAWIREDKTNVDRVLAVTSAVSNQMF 309
YKP+ V GEMRS SLDVWHLAD+Y+ LP LSD+WIREDK N+DRVLAVTSAVSNQ F
Sbjct 478 YKPNMVTGEMRSAYAQSLDVWHLADDYSTLPSLSDSWIREDKANIDRVLAVTSAVSNQFF 537
Query 310 ADLYIQCKATRPMPMYSIPGLIDHH 334
AD+Y++ TRPMPMYS+PGLIDHH
Sbjct 538 ADIYVKNYCTRPMPMYSVPGLIDHH 562
> Gokush_Human_gut_31_045_Microviridae_AG0117_putative.VP1
Length=562
Score = 414 bits (1063), Expect = 1e-142, Method: Compositional matrix adjust.
Identities = 192/265 (72%), Positives = 222/265 (84%), Gaps = 0/265 (0%)
Query 70 NLWAIQSGNVTAATINQLRMAFQIQKLYEKDARGGTRYIEILKSHFGVTSPDARLQRPEY 129
NLWA+ GN AATINQLR+AFQIQK YE+ ARGG+RY E+++S FGVTSPDARLQRPEY
Sbjct 298 NLWAVADGNAAAATINQLRLAFQIQKFYERQARGGSRYTEVVRSFFGVTSPDARLQRPEY 357
Query 130 LGGNRIPVNINQVVQSSATQSSGTPLGDTAAFSVTTDVHGDFIKSFVEHGFVIGIMVARY 189
LGGNR+P+N+NQ+VQ S TQS TP G S+TTD H DF KSF EHG +IG+MVARY
Sbjct 358 LGGNRVPINVNQIVQQSGTQSGTTPQGTVVGQSLTTDKHSDFTKSFTEHGLIIGVMVARY 417
Query 190 DHTYQQGLERFWSRRDRLDYYFPVFANIGEQPILNKEIYAQGTVQDNEVFGYQEAWADYR 249
DHTYQQGL R WSR+D+ D+Y+PVFANIGEQ I NKEI+AQG +DNEVFGYQEAWA+YR
Sbjct 418 DHTYQQGLNRLWSRKDKFDFYWPVFANIGEQAIKNKEIFAQGNDKDNEVFGYQEAWAEYR 477
Query 250 YKPSRVAGEMRSKAPTSLDVWHLADEYTQLPKLSDAWIREDKTNVDRVLAVTSAVSNQMF 309
YKP+ V GEMRS SLDVWHLAD+Y+ LP LSD+WIREDK N+DRVLAVTSAVSNQ F
Sbjct 478 YKPNMVTGEMRSAYAQSLDVWHLADDYSTLPSLSDSWIREDKANIDRVLAVTSAVSNQFF 537
Query 310 ADLYIQCKATRPMPMYSIPGLIDHH 334
AD+Y++ TRPMPMYS+PGLIDHH
Sbjct 538 ADIYVKNYCTRPMPMYSVPGLIDHH 562
> Gokush_Human_gut_27_035_Microviridae_AG0167_putative.VP1
Length=562
Score = 414 bits (1063), Expect = 1e-142, Method: Compositional matrix adjust.
Identities = 192/265 (72%), Positives = 222/265 (84%), Gaps = 0/265 (0%)
Query 70 NLWAIQSGNVTAATINQLRMAFQIQKLYEKDARGGTRYIEILKSHFGVTSPDARLQRPEY 129
NLWA+ GN AATINQLR+AFQIQK YE+ ARGG+RY E+++S FGVTSPDARLQRPEY
Sbjct 298 NLWAVADGNAAAATINQLRLAFQIQKFYERQARGGSRYTEVVRSFFGVTSPDARLQRPEY 357
Query 130 LGGNRIPVNINQVVQSSATQSSGTPLGDTAAFSVTTDVHGDFIKSFVEHGFVIGIMVARY 189
LGGNR+P+N+NQ+VQ S TQS TP G S+TTD H DF KSF EHG +IG+MVARY
Sbjct 358 LGGNRVPINVNQIVQQSGTQSGTTPQGTVVGQSLTTDKHSDFTKSFTEHGLIIGVMVARY 417
Query 190 DHTYQQGLERFWSRRDRLDYYFPVFANIGEQPILNKEIYAQGTVQDNEVFGYQEAWADYR 249
DHTYQQGL R WSR+D+ D+Y+PVFANIGEQ I NKEI+AQG +DNEVFGYQEAWA+YR
Sbjct 418 DHTYQQGLNRLWSRKDKFDFYWPVFANIGEQAIKNKEIFAQGNDKDNEVFGYQEAWAEYR 477
Query 250 YKPSRVAGEMRSKAPTSLDVWHLADEYTQLPKLSDAWIREDKTNVDRVLAVTSAVSNQMF 309
YKP+ V GEMRS SLDVWHLAD+Y+ LP LSD+WIREDK N+DRVLAVTSAVSNQ F
Sbjct 478 YKPNMVTGEMRSAYAQSLDVWHLADDYSTLPSLSDSWIREDKANIDRVLAVTSAVSNQFF 537
Query 310 ADLYIQCKATRPMPMYSIPGLIDHH 334
AD+Y++ TRPMPMYS+PGLIDHH
Sbjct 538 ADIYVKNYCTRPMPMYSVPGLIDHH 562
> Gokush_Human_feces_A_013_Microviridae_AG009_putative.VP1
Length=587
Score = 387 bits (995), Expect = 4e-132, Method: Compositional matrix adjust.
Identities = 178/266 (67%), Positives = 218/266 (82%), Gaps = 2/266 (1%)
Query 71 LWAIQSGNVTAATINQLRMAFQIQKLYEKDARGGTRYIEILKSHFGVTSPDARLQRPEYL 130
L A+ G V+ ATINQLR+AFQIQK YE+ ARGG+RY E+++S FGVTSPDARLQRPEYL
Sbjct 322 LVALNDGAVSVATINQLRLAFQIQKFYERQARGGSRYTEVIRSFFGVTSPDARLQRPEYL 381
Query 131 GGNRIPVNINQVVQ--SSATQSSGTPLGDTAAFSVTTDVHGDFIKSFVEHGFVIGIMVAR 188
GGNR+P+NINQV+Q + ++SS TP G S TTD + DF KSF EHGF+IG+MVAR
Sbjct 382 GGNRVPININQVIQQSGTGSESSSTPQGTVVGMSQTTDTNSDFTKSFTEHGFIIGVMVAR 441
Query 189 YDHTYQQGLERFWSRRDRLDYYFPVFANIGEQPILNKEIYAQGTVQDNEVFGYQEAWADY 248
YDHTYQQGL+R WSR+D+ D+Y+PVFANIGEQ I NKE+YAQGT +D+EVFGYQEAWA+Y
Sbjct 442 YDHTYQQGLDRLWSRKDKFDFYWPVFANIGEQAIKNKELYAQGTAEDDEVFGYQEAWAEY 501
Query 249 RYKPSRVAGEMRSKAPTSLDVWHLADEYTQLPKLSDAWIREDKTNVDRVLAVTSAVSNQM 308
RYKP+RV GEMRS SLD+WHLAD+Y++LP LS WI+ED + V+RVLA + ++ Q
Sbjct 502 RYKPNRVTGEMRSSYAKSLDIWHLADDYSKLPSLSAEWIQEDSSTVNRVLAASDNLAAQF 561
Query 309 FADLYIQCKATRPMPMYSIPGLIDHH 334
FAD+Y++ TRPMPMYSIPGLIDHH
Sbjct 562 FADIYVKNLCTRPMPMYSIPGLIDHH 587
> Gokush_Human_feces_D_014_Microviridae_AG029_putative.VP1
Length=578
Score = 358 bits (920), Expect = 6e-121, Method: Compositional matrix adjust.
Identities = 168/259 (65%), Positives = 201/259 (78%), Gaps = 2/259 (1%)
Query 78 NVTAATINQLRMAFQIQKLYEKDARGGTRYIEILKSHFGVTSPDARLQRPEYLGGNRIPV 137
+ TAATIN+LR+AFQ+QKLYE+DARGGTRYIEI+KSHFGVTSPDARLQRPEYLGG RIP+
Sbjct 320 HATAATINELRLAFQLQKLYERDARGGTRYIEIIKSHFGVTSPDARLQRPEYLGGERIPI 379
Query 138 NINQVVQSSATQSSGTPLGDTAAFSVTTDVHGDFIKSFVEHGFVIGIMVARYDHTYQQGL 197
NI+QV+Q+S T TP G+T A+S+T F SFVEHG+V+G+ R +HTYQQGL
Sbjct 380 NIDQVIQTSGTAEGTTPQGNTGAYSLTGSQGSYFKHSFVEHGYVLGLACVRTEHTYQQGL 439
Query 198 ERFWSRRDRLDYYFPVFANIGEQPILNKEIYAQGTVQDN-EVFGYQEAWADYRYKPSRVA 256
E+ W+R++R D+Y+P ANIGEQ ILNKEIY Q + N E FGYQEAWA+YRYKPSRV+
Sbjct 440 EKIWNRKNRFDFYWPALANIGEQAILNKEIYLQASKATNEEAFGYQEAWAEYRYKPSRVS 499
Query 257 GEMRSKAPT-SLDVWHLADEYTQLPKLSDAWIREDKTNVDRVLAVTSAVSNQMFADLYIQ 315
RS T SLD WH AD Y +LPKLS WI+E NVDR LAV S + +Q AD + +
Sbjct 500 SAFRSNIETGSLDAWHYADYYEELPKLSAEWIQETYKNVDRTLAVQSTLEDQYIADFWFK 559
Query 316 CKATRPMPMYSIPGLIDHH 334
CK TRPMP+YSIPGLIDHH
Sbjct 560 CKCTRPMPIYSIPGLIDHH 578
> Gokush_Human_feces_B_029_Microviridae_AG0417_putative.VP1
Length=530
Score = 338 bits (868), Expect = 9e-114, Method: Compositional matrix adjust.
Identities = 161/256 (63%), Positives = 192/256 (75%), Gaps = 3/256 (1%)
Query 79 VTAATINQLRMAFQIQKLYEKDARGGTRYIEILKSHFGVTSPDARLQRPEYLGGNRIPVN 138
VT+ATINQLR AFQIQKL EKDARGGTRY E+L+ HFGV SPD+R+Q PEYLGG R+P+N
Sbjct 278 VTSATINQLRQAFQIQKLLEKDARGGTRYREVLREHFGVISPDSRMQIPEYLGGYRLPIN 337
Query 139 INQVVQSSATQSSGTPLGDTAAFSVTTDVHGDFIKSFVEHGFVIGIMVARYDHTYQQGLE 198
++QV+Q+S+T S+ +PLG+TAA SVTT F KSF EHGF++G+ V R D TYQQG+E
Sbjct 338 VSQVIQTSSTDST-SPLGNTAALSVTTMNKPMFTKSFTEHGFIMGLAVVRTDQTYQQGIE 396
Query 199 RFWSRRDRLDYYFPVFANIGEQPILNKEIYAQGTVQDNEVFGYQEAWADYRYKPSRVAGE 258
R WSR+ R DYY+PV ANIGEQ ILNKEIYAQG+ +D E FGYQEAWADYRYKPS+V
Sbjct 397 RMWSRKGRYDYYWPVLANIGEQAILNKEIYAQGSAKDEEAFGYQEAWADYRYKPSKVTAL 456
Query 259 MRSKAPTSLDVWHLADEYTQLPKLSDAWIREDKTNVDRVLAVTSAVSNQMFADLYIQCKA 318
RS A SLD WH A +Y +LP LS AW+ + + R LA + AD Y K
Sbjct 457 FRSNAQQSLDAWHYAQDYNELPTLSTAWMEQSNAEMKRTLARSD--QPDFIADFYFMNKT 514
Query 319 TRPMPMYSIPGLIDHH 334
TR MP+YSIPGLIDHH
Sbjct 515 TRCMPVYSIPGLIDHH 530
> Gokush_JCVI_001_Microviridae_AG025_putative.VP1
Length=560
Score = 313 bits (803), Expect = 1e-103, Method: Compositional matrix adjust.
Identities = 161/295 (55%), Positives = 204/295 (69%), Gaps = 13/295 (4%)
Query 42 LGGTDVESVATGVGFDAPTSRDGSMYLDNLWAIQSGNVTAATINQLRMAFQIQKLYEKDA 101
+GGT V+ V +G TS D +Y D A TAATINQLR +FQIQ+L E+DA
Sbjct 275 VGGT-VQGVDKALG--VATSGDTGIYADLSAA------TAATINQLRESFQIQRLLERDA 325
Query 102 RGGTRYIEILKSHFGVTSPDARLQRPEYLGGNRIPVNINQVVQSSATQS--SGTPLGDTA 159
RGGTRY EI++SHFGV SPDARLQRPEYLGG PVNI+ + Q+SAT S + TPLG+ A
Sbjct 326 RGGTRYTEIIRSHFGVISPDARLQRPEYLGGGSTPVNISPIAQTSATASGATATPLGNLA 385
Query 160 AFSVTTDVHGDFIKSFVEHGFVIGIMVARYDHTYQQGLERFWSRRDRLDYYFPVFANIGE 219
A + F +SFVEHG VIG++ R D TYQQG+ + WSRR R D+YFPVFA++GE
Sbjct 386 AMGTALAMGHGFTQSFVEHGHVIGLVAVRADLTYQQGMRKMWSRRTRYDFYFPVFAHLGE 445
Query 220 QPILNKEIYAQGTVQDNEVFGYQEAWADYRYKPSRVAGEMRSKAPTSLDVWHLADEYTQL 279
Q +LNKEIY GT D++VFGYQE WA+YRY PS++ RS A +LD WHLA ++ L
Sbjct 446 QAVLNKEIYTTGTSTDDDVFGYQERWAEYRYHPSQITSLFRSTAAGTLDAWHLAQNFSSL 505
Query 280 PKLSDAWIREDKTNVDRVLAVTSAVSNQMFA-DLYIQCKATRPMPMYSIPGLIDH 333
P L+ ++I +D VDRV+A+ + + Q F D + KA RPMP+YS+PGLIDH
Sbjct 506 PTLNTSFI-QDNPPVDRVVAIGAEANGQQFIFDSFFDIKAARPMPLYSVPGLIDH 559
Lambda K H a alpha
0.318 0.133 0.404 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 28557990