bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-1_CDS_annotation_glimmer3.pl_2_4
Length=566
Score E
Sequences producing significant alignments: (Bits) Value
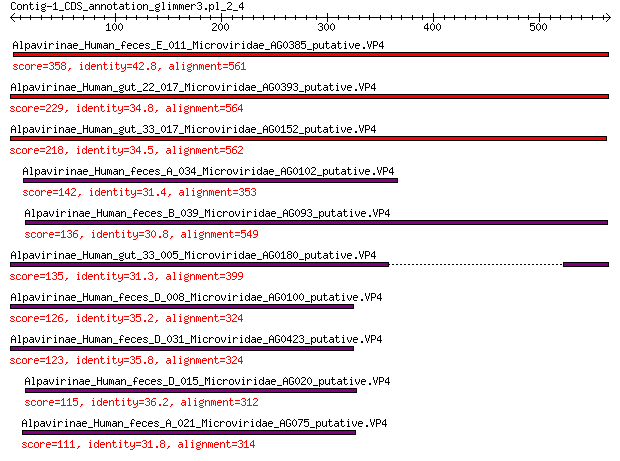
Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4 358 5e-117
Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4 229 1e-68
Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4 218 1e-64
Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4 142 2e-38
Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4 136 3e-36
Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4 135 6e-36
Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4 126 6e-33
Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4 123 6e-32
Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4 115 7e-29
Alpavirinae_Human_feces_A_021_Microviridae_AG075_putative.VP4 111 9e-28
> Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4
Length=611
Score = 358 bits (919), Expect = 5e-117, Method: Compositional matrix adjust.
Identities = 240/627 (38%), Positives = 337/627 (54%), Gaps = 89/627 (14%)
Query 4 QEVIDKYLLAECLHPVKVLNKYTNDFIYSPCGHCYSCLKNKSNRDTALAMNIASNFKYCY 63
+E++ KYL ECL P +++N Y++D I++PCGHC SC+ NKSN TA AMN+A++FKYCY
Sbjct 5 EELLTKYLFTECLRPQRIVNPYSHDVIFAPCGHCKSCIMNKSNFATAYAMNMATHFKYCY 64
Query 64 FVWLSYEDQYLPYMELK-------------------------------------SIDSLD 86
FV L+Y+D +LPY+ ++ S+DS
Sbjct 65 FVTLTYKDIFLPYLSVEVVRRSGNRYLFDENFETMVSTSDPRFLTPEYYHDRDFSLDSAQ 124
Query 87 NNRSNYF---FSSVNRNLRILVPNG-KDRIIEDPLFEFTHPMTSFEYQDIIVKSHGRYDF 142
N F F S+ R++ + + R +D +F PM E QDI++K++GRYD+
Sbjct 125 NEVEQVFDVGFQSIPRDVSVKSKGSFRFRSFDDEPLKFCIPMKLTELQDILIKANGRYDY 184
Query 143 LRKCVVYPRFEDCDNRIPYCNTSDCQKFLKRLR--FHSKNKYNEEIRFYGVSEYGPRTYR 200
+K VVYP DC +IP + D + F KRLR S + +I +Y VSEYGP+TYR
Sbjct 185 GKKKVVYPSLADCKLQIPVLQSRDIELFFKRLRRNLDSHGFTSSKICYYVVSEYGPQTYR 244
Query 201 PHWHLLLFFNSDELTSVIQQFVSESWSYGRTTCELsrggsssyvasyvnsnVCLPSLYLQ 260
PHWH LLFFNS+E+T +++ +S++WSYGR LSRG ++SYVA+YVNS CLP LY+
Sbjct 245 PHWHCLLFFNSEEITQTLREDISKAWSYGRIDYSLSRGAAASYVAAYVNSAACLPFLYVG 304
Query 261 HKEIRARSLHSKGYGNNYVFPTQATIHEFDKMYSLLLNGESISFNGKAKQIYPSRSYKHT 320
KEIR RS HSKG+G+N VFP + + E K+ L +G ++ NGK I P RS +
Sbjct 305 QKEIRPRSFHSKGFGSNKVFPKSSDVSEISKISDLFFDGVNVDSNGKVVNIRPVRSCELA 364
Query 321 VFPRFSNLVCKsahssafvfsaaffTPERLVRYGYLDITYDKSVSP---VSQLAHAYTDF 377
VFPRFSN + + +F + TPERLV GYL I S +S L AY+++
Sbjct 365 VFPRFSNDFFSDSDTCCKLFQSVIETPERLVSRGYLGIDTPNFGSDDFRLSDLVRAYSEY 424
Query 378 FLNR-----------EDKRSISFDDELIITTVRL-DAPNRKYWHCLTYDQIYSKFYRLFN 425
+ R+ + DELI RL D + D I+ + YRLF
Sbjct 425 YERNFTDFSFSLRFLRGYRTRDYADELIFREARLFDG------YVFNKDMIFGRLYRLFA 478
Query 426 TVIRTARFWNLFAYVDSCR-RGPIYDLMNFSDNYWNEFARRRLSDYFAFLENCNDRQRSF 484
V+R RFWNL Y DS + I + ++ YW + R L+ YF +LE CND +R F
Sbjct 479 KVLRCFRFWNLKQYTDSWSLKVAIKKIWSYGWEYWKKKEYRFLTTYFEYLEGCNDDERLF 538
Query 485 LFHHTALNEVFNSRFVSRFKTVDKGGYELTESFSEFDKRDYMYDSTLSQEF-------LK 537
L T+ + + + S++ + DY+ + LS +F LK
Sbjct 539 LLVRTSGSGL---------------ATDSPHSWTYTQREDYV--NCLSDDFYKRYMNTLK 581
Query 538 ELTAINRKACVDKVKHKEFNDLSGLLL 564
LTA K DKVKHKEFND+ G+LL
Sbjct 582 WLTARTEKVLKDKVKHKEFNDMQGVLL 608
> Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4
Length=562
Score = 229 bits (583), Expect = 1e-68, Method: Compositional matrix adjust.
Identities = 196/608 (32%), Positives = 297/608 (49%), Gaps = 90/608 (15%)
Query 1 MINQEVIDKYLLAECLHPVKVLNKYTNDFIYSPCGHCYSCLKNKSNRDTALAMNIASNFK 60
MI +E+ +K L+ C HP V+NKYT++ + CGHC SC+ +S+ T L ++ F+
Sbjct 1 MITKELQNK-LVTRCQHPRTVVNKYTHEPVVVSCGHCPSCILRRSSVQTNLLTTYSAQFR 59
Query 61 YCYFVWLSYEDQYLPYMELK----------------SIDSLDNNRSNYF---FSSVNRNL 101
Y YFV L+Y +LP +E+ +ID LD + N + F SV R+
Sbjct 60 YVYFVTLTYAPSFLPTLEVSVVETCTDDIADVSCVPNIDELDASDPNTYLFGFRSVPRSS 119
Query 102 RI-LVPNGKDRIIEDPLFEFTHPMTSFEYQDIIVKSHGRYDFLRKCVVYPRFEDCDNRIP 160
+ L + +R +DP +F++PM E I+ K + + NRIP
Sbjct 120 SVKLKSSTVERTFKDPDVKFSYPMKPKELLSILGKIN---------------HNVPNRIP 164
Query 161 YCNTSDCQKFLKRLRFHSKNKYNEEIRFYGVSEYGPRTYRPHWHLLLFFNSDELTSVIQQ 220
Y D FLKRLR + +E++R+Y VSEYGP ++RPHWHLLLF NS+ + + +
Sbjct 165 YICNRDLDLFLKRLRSY---YLDEKLRYYAVSEYGPTSFRPHWHLLLFSNSERFSRTVCE 221
Query 221 FVSESWSYGRTTCELsrggsssyvasyvnsnVCLPSLYLQH-KEIRARSLHSKGYGNNYV 279
VS++WSYGR LSRG ++ YVASYVNS V LP Y Q K +R +S HS G+ + +
Sbjct 222 NVSKAWSYGRCDASLSRGFAAPYVASYVNSFVALPDFYTQMPKVVRPKSFHSIGFTESNL 281
Query 280 FPTQATIHEFDKMYSLLLNGESISFNGKAKQIYPSRSYKHTVFPRFSNLVCKsahssafv 339
FP + + E D++ LNG + +G + I P+ Y +FPRFS+ + KS S +
Sbjct 282 FPRKVRVAEVDEITDKCLNGVRVERDGYFRTIKPTWPYLLRLFPRFSDAIRKSPSSIYQL 341
Query 340 fsaaffTPERLVRYGYLDITYDKSVSPVSQLAHAYTDFFLNRED--------------KR 385
AAF PER++R G DI D Q ++ +LN D +
Sbjct 342 LFAAFTAPERVIRSGCADIGCDPFGESSKQSILSFCKQYLNYVDNYGKSNEHRNFLSPQA 401
Query 386 SISFDDELIITTVRL-DAPNRKYWHCLTYDQIY------SKFYRLFNTVIRTARFWNLFA 438
S+ D LI++ RL D + + H ++ ++Y SKF R ++T + FW+
Sbjct 402 SLPHSDVLILSECRLYDGVDLETTHRVS--RVYRFFLGISKFIRTYSTDGCSELFWSSGT 459
Query 439 YVDS--CRRGPIYDLMNFSDNYWNEFARRRLSDYFAFLENCNDRQRSFLFHHTALNEVFN 496
CR + + N+WN + RL D++ LE+ ND++ L N F
Sbjct 460 PGGDLFCRERFLRIISEKIVNFWNRYEYNRLVDFYQTLEDSNDKE---LVDFELRNYSF- 515
Query 497 SRFVSRFKTVDKGGYELTESFSEFDKRDYMYDSTLSQEFLKELTAINRKACVDKVKHKEF 556
R+ + ++ EL ++ L A + C DKVKHK+
Sbjct 516 -RYNRSVRDNERPYNEL--------------------PLVRRLAAASLMKCRDKVKHKKV 554
Query 557 NDLSGLLL 564
NDLSG+ L
Sbjct 555 NDLSGIFL 562
> Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4
Length=565
Score = 218 bits (556), Expect = 1e-64, Method: Compositional matrix adjust.
Identities = 194/611 (32%), Positives = 289/611 (47%), Gaps = 100/611 (16%)
Query 1 MINQEVIDKYLLAECLHPVKVLNKYTNDFIYSPCGHCYSCLKNKSNRDTALAMNIASNFK 60
MI +E+ +K L+ C +P V+NKYT++ + CG C SC+ +S T L ++ F+
Sbjct 1 MITKELQNK-LVTRCQNPRTVVNKYTHEPVVVSCGACPSCVLRRSGIQTNLLTTYSAQFR 59
Query 61 YCYFVWLSYEDQYLPYMELKSI-------------------DSLDNNRSNYFFSSVNRNL 101
Y YFV L+Y +LP +E+ I D DNNR + F SV R+
Sbjct 60 YVYFVTLTYAPCFLPTLEVSVIETCTDDIADVSSVPDINDLDPCDNNRYLFGFCSVPRSA 119
Query 102 RILVPNGK-DRIIEDPLFEFTHPMTSFEYQDIIVKSHGRYDFLRKCVVYPRFEDCDNRIP 160
+ + N +R +DP F++PM E I+ K + + NRIP
Sbjct 120 SVKLKNSTVERTFKDPEVRFSYPMKPKELLSILDKIN---------------HNVPNRIP 164
Query 161 YCNTSDCQKFLKRLRFHSKNKYNEEIRFYGVSEYGPRTYRPHWHLLLFFNSDELTSVIQQ 220
Y D FLKRLR + E++R+Y VSEYGP +YRPHWHLLLF NS++ + I +
Sbjct 165 YICNRDLDLFLKRLRSYYPY---EKLRYYAVSEYGPTSYRPHWHLLLFSNSEQFSKTILE 221
Query 221 FVSESWSYGRTTCELsrggsssyvasyvnsnVCLPSLYLQHKE-IRARSLHSKGYGNNYV 279
VS++WSYGR LSRG ++ YVASYVNS V LPS Y + +R +S HS G+ + +
Sbjct 222 NVSKAWSYGRCDASLSRGFAAPYVASYVNSFVALPSFYTEMPRFLRPKSFHSIGFTESNL 281
Query 280 FPTQATIHEFDKMYSLLLNGESISFNGKAKQIYPSRSYKHTVFPRFSNLVCKsahssafv 339
FP + I E D++ LNG + +G + + PS Y +FPRFS+ + KS S +
Sbjct 282 FPRKVRISEIDEVTDKCLNGVRVERDGYFRILKPSWPYLLRLFPRFSDAIRKSPSSIYQL 341
Query 340 fsaaffTPERLVRYGYLDITYDKSVSPVSQLAHAYTDFFLNREDKRS------------- 386
AAF P R++R G DI D V Q ++ +LN D
Sbjct 342 LFAAFTAPARVIRSGCADIGCDPFVENSKQSLLSFCKQYLNYVDNHGKSNEFKNFLSPHA 401
Query 387 -ISFDDELIITTVRL-DAPNRKYWHCLTYDQIY------SKFYRLFNTVIRTARFWNLFA 438
+ D LI+T RL D + + H L+ ++Y +KF R ++T + FW+
Sbjct 402 DLPHSDVLILTECRLYDGVDLEAAHRLS--RVYRFFLGIAKFIRTYSTDGCSELFWS--- 456
Query 439 YVDSCRRGPIYDLMNF----SDN---YWNEFARRRLSDYFAFLENCNDRQRSFLFHHTAL 491
G ++ F S+ +WN + RL D++ LE+ ND++
Sbjct 457 --SGTPGGELFGRERFLRIISEKIVVFWNRYDYMRLVDFYQTLEDSNDKELVDF------ 508
Query 492 NEVFNSRFVSRFKTVDKGGYELTESFSEFDKRDYMYDSTLSQEFLKELTAINRKACVDKV 551
E+ N F DK + + E ++ L A + C DKV
Sbjct 509 -EIRNYSFCYNRSVRDK-----EKPYHEL-------------PLVRRLAAASLMKCRDKV 549
Query 552 KHKEFNDLSGL 562
KHK+ ND+ G+
Sbjct 550 KHKKINDMFGI 560
> Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4
Length=539
Score = 142 bits (358), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 111/362 (31%), Positives = 178/362 (49%), Gaps = 20/362 (6%)
Query 13 AECLHPVKVLNKYTNDFIYSPCGHCYSCLKNKSNRDTALAMNIA-SNFKYCYFVWLSYED 71
CLHP + NKYT D IY PCG C C+ NK+ + L N+ ++ +YC F+ L+Y
Sbjct 10 GSCLHPRVIKNKYTGDPIYVPCGTCEFCVHNKAIK-AELKCNVQLASSRYCEFITLTYST 68
Query 72 QYLPYMELKSIDSLDNNRSNYFFSSVNRNL----RILVPNGKDRIIEDPLFEFTHPMTSF 127
+YLP + R F S+ R+ + + ++ D F+F ++
Sbjct 69 EYLPVGKF-----YQGPRGEVRFCSLPRDFVYSYKTVQGYTRNISFNDKTFDFDTQLSWS 123
Query 128 EYQDIIVKSHGRY-DFLRKCVVYPRFEDCDNRIPYCNTSDCQKFLKRLRFHSKNKYNEEI 186
Q + K+H Y F +Y R + + Y N D Q F KRL + + NE+I
Sbjct 124 SAQLLQKKAHLHYTSFPDGRCIYNR-PYMEGLVGYLNYHDIQLFFKRLNQNIRRITNEKI 182
Query 187 RFYGVSEYGPRTYRPHWHLLLFFNSDELTSVIQQFVSESWSYGRTTCELsrggsssyvas 246
+Y V EYGP T+RPH+H+LLF +S +L I+QFVS+SW +G + + +S YVA
Sbjct 183 YYYVVGEYGPTTFRPHFHILLFHDSRKLRESIRQFVSKSWRFGDSDTQPVWSSASCYVAG 242
Query 247 yvnsnVCLPSLYLQHKEIRARSLHSKGYGN---NYVFPTQATIHEFDKMYSLLLNGESIS 303
YVNS CLP + + I+ S + N VF + E ++++SL +G ++
Sbjct 243 YVNSTACLPDFFKNSRHIKPFGRFSVNFAESAFNEVFKPE----ENEEIFSLFYDGRVLN 298
Query 304 FNGKAKQIYPSRSYKHTVFPRFSNLVCKsahssafvfsaaffTPERLVRYGYLDITYDKS 363
NGK + P RS+ + ++PR + + S P+ + ++G++D D
Sbjct 299 LNGKPTIVRPKRSHINRLYPRLDKSKHATVVDDIRIASVVSRLPQVVAKFGFIDEVSDFE 358
Query 364 VS 365
+S
Sbjct 359 LS 360
> Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4
Length=559
Score = 136 bits (342), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 169/599 (28%), Positives = 259/599 (43%), Gaps = 107/599 (18%)
Query 15 CLHPVKVLNKYTNDFIYSPCGHCYSCLKNKSNRDTALAMNIASNFKYCYFVWLSYEDQYL 74
C H + N+Y I CG C C+ ++ + + S FKY YFV L+Y+++++
Sbjct 14 CQHRSFITNRYNGARITVDCGQCDYCIHKRAQKASMRVKTAGSAFKYSYFVTLTYDNEHI 73
Query 75 PYMELKSIDS----------------------------LDNNR--SNYFFSSV------N 98
P M K + S D+N + FF V +
Sbjct 74 PLMNCKVLHSEYEDVVGISGDIHFGDEYHKYIPVSEYRCDDNSMLRHIFFEQVQGTVPFD 133
Query 99 RNLRILVPNGKDRIIEDPLFEFTHPMTSFEYQDIIVKSHGRYDFLRKCVVYPRFEDC--D 156
R ++ VP ++D F + SF Y+ +S + D YP E D
Sbjct 134 REIKEYVP------VKDNWFLSMAAIRSFIYK---TQSVDKTD-------YPASEQYGRD 177
Query 157 NRIPYCNTSDCQKFLKRLR---FHSKNKYNEEIRFYGVSEYGPRTYRPHWHLLLFFNSDE 213
N IP+ N D Q ++KRLR F KY E FY V EYGP +RPH+HLLLF NSD+
Sbjct 178 NLIPFLNYVDVQNYIKRLRKYLFQQLGKY-ETFHFYAVGEYGPVHFRPHYHLLLFTNSDK 236
Query 214 LTSVIQQFVSESWSYGRTTCELsrggsssyvasyvnsnVCLPSLYLQHKEIRARSLHSKG 273
++ V++ +SW GR+ + S GG+ SYVASYVNS P LY + R +S S G
Sbjct 237 VSEVLRYCHDKSWKLGRSDFQRSAGGAGSYVASYVNSLCSAPLLYRSCRAFRPKSRASVG 296
Query 274 Y---GNNYVFPTQATIHEFDKMYSLLLNGESISFNGKAKQIYPSRSYKHTVFPRFSNLVC 330
+ G ++V + ++ ++NG +FNG + + P SY T+ PRFS+
Sbjct 297 FFEKGCDFV-EDDDPYAQIEQKIDSVVNGRVYNFNGVSVRSTPPLSYIRTLLPRFSSARN 355
Query 331 KsahssafvfsaaffTPERLVRYGYLDITYDKSVSPVSQLAHAYTDFFLNREDKRSISFD 390
+ A + A TP+R+ R+G+ D D +S L Y + + ++ D
Sbjct 356 DDGIAIARILYAVHSTPKRIARFGFADYKQDSILS----LVRTYYQYL---KVNPILTDD 408
Query 391 DELIITTVRLDAPNRKYWHCLTYDQIYS---KFYRLFNTVIRTARFWNL--FAYVDSCRR 445
D+LI+ R ++ +C + I S K YRLF V + R W+L F S
Sbjct 409 DKLILHASRCLT---RFVNCSSDVDIESYINKLYRLFLYVYKFFRNWHLPFFGSDISAFS 465
Query 446 GPIYDLMNFSDNYWNEFARRRLSDYFAFLENCNDRQRSFLFHHTALNEVFNSRFVSRFKT 505
G I ++ Y + DY +L +VFN R S
Sbjct 466 GRIMFIIKTGIEY------EKKKDY------------------ESLRDVFNIR--SANPN 499
Query 506 VDKGGYELTESFSEFDKRDYMYD-STLSQEFLKELTAINRKACVDKVKHKEFNDLSGLL 563
+ + L + E RD + D S + + L++L + C D +KHK ND + +
Sbjct 500 ISDCMFALPANGQE---RDVLSDVSCETVQLLEQLRLRSATFCRDMIKHKRLNDANNIF 555
> Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4
Length=536
Score = 135 bits (340), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 112/365 (31%), Positives = 174/365 (48%), Gaps = 22/365 (6%)
Query 1 MINQEVIDKYLLAECLHPVKVLNKYTNDFIYSPCGHCYSCLKNKS-NRDTALAMNIASNF 59
MIN V+ Y CLHPV + NKYT D IY CG C CL NK+ ++ + +AS+
Sbjct 1 MIN-PVVKPY--CSCLHPVIIKNKYTGDPIYVECGTCEVCLSNKAIQKELRCNIQLASS- 56
Query 60 KYCYFVWLSYEDQYLPYMELKSIDSLDNNRSNYFFSSVNRN--LRILVPNGKDRIIE--D 115
+ C+FV L+Y +++P ++ +Y V R+ + G +R + D
Sbjct 57 RCCFFVTLTYATEHIPVARFYKLND------SYHLCCVPRDHVYTYVTSQGYNRKMSFCD 110
Query 116 PLFEFTHPMTSFEYQDIIVKSH---GRYDFLRKCVVYPRFEDCDNRIPYCNTSDCQKFLK 172
F++ + ++ K+H Y R V YP D +PY N D Q F K
Sbjct 111 EEFDYPTNLRDEAVTALLDKTHLDRTVYPDGRSVVKYPNMGDL---LPYLNYRDVQLFHK 167
Query 173 RLRFHSKNKYNEEIRFYGVSEYGPRTYRPHWHLLLFFNSDELTSVIQQFVSESWSYGRTT 232
R+ K +E+I Y V EYGP+T+RPH+HLL FF+S+ L V Q V ++W +G +
Sbjct 168 RINQQIKKYTDEKIYSYTVGEYGPKTFRPHFHLLFFFDSERLAQVFGQLVDKAWRFGNSD 227
Query 233 CELsrggsssyvasyvnsnVCLPSLYLQHKEIRARSLHSKGYGNNYVFPTQATIHEFDKM 292
+ +SSYVA Y+NS+ CLP Y +++I S+ + F E +K+
Sbjct 228 TQRVWSSASSYVAGYLNSSHCLPEFYRCNRKIAPFGRFSQYFAER-PFIEAFKPEENEKV 286
Query 293 YSLLLNGESISFNGKAKQIYPSRSYKHTVFPRFSNLVCKsahssafvfsaaffTPERLVR 352
+ +NG +S GK P RS + ++P S+ P+ L +
Sbjct 287 FDKFVNGIYLSLGGKPTLCRPKRSLINRLYPVLDRSAVSDVDSNVRTALFVSKIPQVLAK 346
Query 353 YGYLD 357
+G+LD
Sbjct 347 FGFLD 351
Score = 23.1 bits (48), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 13/42 (31%), Positives = 23/42 (55%), Gaps = 4/42 (10%)
Query 523 RDYMYDSTLSQEFLKELTAINRKACVDKVKHKEFNDLSGLLL 564
R+ + S+ + E+ A NR+ ++KHK NDL+ L+
Sbjct 493 REVLALSSFCRAAYAEVAADNRQ----RIKHKYLNDLNCALI 530
> Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4
Length=532
Score = 126 bits (317), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 114/344 (33%), Positives = 170/344 (49%), Gaps = 43/344 (13%)
Query 1 MINQEVIDKYLLAECLHPVKVLNKYTNDFIYSPCGHCYSCLKNKS--NRDTALAMNIASN 58
M QE I K + + C H V+ N YT + I PCG C +C NKS +++ A ++ S
Sbjct 1 MNQQEFIQK-VFSMCQHQVQTKNPYTGELIMVPCGTCPACRFNKSILSQNKVHAQSLIS- 58
Query 59 FKYCYFVWLSYEDQYLPYMELKSIDSLDNN--------------RSNYFFSSVNRNLRIL 104
++ YFV L+Y +Y+PY E + I++LD + Y + LRI
Sbjct 59 -RHVYFVTLTYAQRYIPYYEYE-IEALDADFLAITAHCRDRNPMYRTYTYRGTKHKLRI- 115
Query 105 VPNGKDRIIEDPLFE-FTHPMTSFEYQDIIVKSHGRYDFLRKCVVYPRFEDCDNRIPYCN 163
R + P + F+ + + K++ +D YP D RIPY
Sbjct 116 ------RGLASPKVKSFSFSVNRDYWTSYAQKANLSFD-----GKYPALSD---RIPYLL 161
Query 164 TSDCQKFLKRLR-FHSKNKYNEEIRFYGVSEYGPRTYRPHWHLLLFFNSDELTSVIQQFV 222
D ++KR+R + SK NE I Y V EYGP T+RPH+HLLLFFNSDEL I +
Sbjct 162 HDDVSLYMKRVRKYISKLGINETIHTYVVGEYGPVTFRPHFHLLLFFNSDELAQSIVRIA 221
Query 223 SESWSYGRTTCELsrggsssyvasyvnsnVCLPSLYLQHKEIRARSLHSKGYGNNYVFPT 282
W +GR C SRG + YV+SY+NS +P + + IR + S +G ++ +
Sbjct 222 RSCWRFGRVDCSASRGDAEDYVSSYLNSFSSIPLHIQEIRAIRPFARFSNKFGFSFFESS 281
Query 283 --QATIHEFDKMYSLLLNGESISFNGKAKQIYPSRSYKHTVFPR 324
+A +FD+ +LNG+S+ +NG I+P R+ T F R
Sbjct 282 IKKAQSGDFDE----ILNGKSLPYNGFNTTIFPWRAIIDTCFYR 321
> Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4
Length=532
Score = 123 bits (309), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 116/349 (33%), Positives = 170/349 (49%), Gaps = 53/349 (15%)
Query 1 MINQEVIDKYLLAECLHPVKVLNKYTNDFIYSPCGHCYSCLKNKS--NRDTALAMNIASN 58
M QE I K + + C H V+ N YT + I PCG C +C NKS +++ A ++ S
Sbjct 1 MNQQEFIQK-VFSMCQHQVQTKNPYTGELIMVPCGTCPACRFNKSILSQNKVHAQSLVS- 58
Query 59 FKYCYFVWLSYEDQYLPYMELKSIDSLDNN--------------RSNYFFSSVNRNLRI- 103
++ YFV L+Y +Y+PY E + I++LD + Y + LRI
Sbjct 59 -RHVYFVTLTYAQRYIPYYEYE-IEALDADFLAITAHCCDRNPMYRTYTYRGTKHKLRIR 116
Query 104 --LVPNGKDRIIEDPLFEFT---HPMTSFEYQDIIVKSHGRYDFLRKCVVYPRFEDCDNR 158
PN K F F+ TS+ Q + +G+Y L R
Sbjct 117 GLASPNVKS-------FSFSVNRDYWTSYA-QKANLSFNGKYPAL------------SGR 156
Query 159 IPYCNTSDCQKFLKRLR-FHSKNKYNEEIRFYGVSEYGPRTYRPHWHLLLFFNSDELTSV 217
IPY D ++KR+R + SK NE I Y V EYGP ++RPH+HLLLFF+SDEL
Sbjct 157 IPYLLHGDVSLYMKRVRKYISKLGINETIHTYIVGEYGPSSFRPHFHLLLFFDSDELAQN 216
Query 218 IQQFVSESWSYGRTTCELsrggsssyvasyvnsnVCLPSLYLQHKEIRARSLHSKGYGNN 277
I + S W +GR C SRG + YV+SY+NS +P + + IR + S +G +
Sbjct 217 IIRIASSCWRFGRVDCSASRGDAEDYVSSYLNSFSSIPLHIQEIRAIRPFARFSNKFGYS 276
Query 278 YVFPT--QATIHEFDKMYSLLLNGESISFNGKAKQIYPSRSYKHTVFPR 324
+ + +A FD+ +LNG+S+ +NG I+P R+ T F R
Sbjct 277 FFESSIKKAQSGNFDE----ILNGKSLPYNGFDTTIFPWRAIIDTCFYR 321
> Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4
Length=563
Score = 115 bits (287), Expect = 7e-29, Method: Compositional matrix adjust.
Identities = 113/378 (30%), Positives = 170/378 (45%), Gaps = 82/378 (22%)
Query 15 CLHPVKVLNKYTNDFIYSPCGHCYSCLKNKSNRDTALAMNIASNFKYCYFVWLS------ 68
C HP + NKYT D++ CG C CL K++R T + N +YCYFV L+
Sbjct 12 CEHPQVIQNKYTGDYVKVDCGQCPYCLIKKADRSTQKCDFVKYNHRYCYFVTLTYNTQYV 71
Query 69 ----------YEDQYLPYMELKSIDS------LDNNRSN-----YFFSSVNR-----NLR 102
Y ++LP KS + L ++R N + + VNR +LR
Sbjct 72 PKMSLTQIEDYLSEWLPVRPPKSFGTQLTARMLTDSRVNKKIPDFMSAEVNRPYMLEHLR 131
Query 103 IL--------------------------VP------NGKDRIIEDPLFEFTHPMTSFEYQ 130
+L +P N KD E+ ++ S + +
Sbjct 132 LLEADRYKALALRYPNFISKARPYILRSIPRVSKLQNFKDEYFEELVWMLPEIAESLKKK 191
Query 131 DIIVKSHGRYDFLRKCVVYPRFEDCDNRIPYCNTSDCQKFLKRLRFHSKNKYN--EEIRF 188
+ ++G +P+F+ + Y N D Q F KRLR + K E+I
Sbjct 192 NN-TDANG---------AFPQFKGL---LKYVNIRDYQLFAKRLRKYLSKKVGKYEKIHS 238
Query 189 YGVSEYGPRTYRPHWHLLLFFNSDELTSVIQQFVSESWSYGRTTCELsrggsssyvasyv 248
Y VSEY P+T+RPH+H+L FF+SDE+ +Q V +SW GR +L+R ++SYV++Y+
Sbjct 239 YVVSEYSPKTFRPHFHILFFFDSDEIAKNFRQAVYQSWRLGRVDTQLAREQANSYVSNYL 298
Query 249 nsnVCLPSLYLQHKEIRARSLHSKGYGNNYVFPTQATIHEFDKMYSLLLNGESISFNGKA 308
NS V +P +Y K IR RS S +G V + I + + L +G S N K
Sbjct 299 NSVVSIPFVYKAKKSIRPRSRFSNLFGFEEV---KEGIRKAQDKRAALFDGLSYISNQKF 355
Query 309 KQIYPSRSYKHTVFPRFS 326
+ PS S +FPRF+
Sbjct 356 VRYVPSGSLIDRLFPRFT 373
> Alpavirinae_Human_feces_A_021_Microviridae_AG075_putative.VP4
Length=547
Score = 111 bits (278), Expect = 9e-28, Method: Compositional matrix adjust.
Identities = 100/330 (30%), Positives = 149/330 (45%), Gaps = 31/330 (9%)
Query 12 LAECLHPVKVLNKYTNDFIYSPCGHCYSCLKNKSNRDTALAMNIASNFKYCYFVWLSYED 71
+++C P + +++D + C C CL KS DT FKY +FV L Y
Sbjct 13 MSDCNQPRYLRQLHSDDVLSIGCNECLPCLLKKSRNDTLDVTISTQTFKYIFFVNLDYAT 72
Query 72 QYLPYMELKSIDSLDNNRSNYFFSSVNRNLRILVPNGKDR--------------IIEDPL 117
++P M + I + Y S V +++I P K R E
Sbjct 73 PFIPIMRHEYIP---ERKVIYCESMVRPSVKI--PTNKRRKDGSVVFTTFSFRAADETRP 127
Query 118 FEFTHPMTSFEYQDIIVKSHGRYDFLRKCVVYPRFEDCDNRIPYCNTSDCQKFLKRLRFH 177
F FT P S E + V+ L YP ++DC PY + D F+KRLR
Sbjct 128 FCFTAPCESEEEYRLFVRKVN----LTANGKYPMYKDC---FPYLSRYDVALFMKRLRNL 180
Query 178 SKNKYNEEIRF--YGVSEYGPRTYRPHWHLLLFFNSDELTSVIQQFVSESWSYGRTTCEL 235
KY + I Y V EYGP +RPH+H+++ N ++ FV ++W +G + E+
Sbjct 181 FLKKYGQSIYMHSYIVGEYGPVHFRPHYHIIISSNDPRFAESLEHFVDKAWFFGGASAEI 240
Query 236 srggsssyvasyvnsnVCLPSLYLQHKEIRARSLHSKGYGNNYVFPTQATIHEFDKMYSL 295
G +SYVASYVNS+V LP L+ +H+ +R S +G+ P F +
Sbjct 241 PDGNCASYVASYVNSSVSLPVLFRKHRLVRPFSRKCRGFLMERFAPDSLP---FGERTDR 297
Query 296 LLNGESISFNGKAKQIYPSRSYKHTVFPRF 325
+NG S + Q+YP RSY+ V+PR
Sbjct 298 DINGTLCSIGDASFQLYPRRSYRDRVYPRL 327
Lambda K H a alpha
0.324 0.139 0.434 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 51994738