bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-17_CDS_annotation_glimmer3.pl_2_6
Length=367
Score E
Sequences producing significant alignments: (Bits) Value
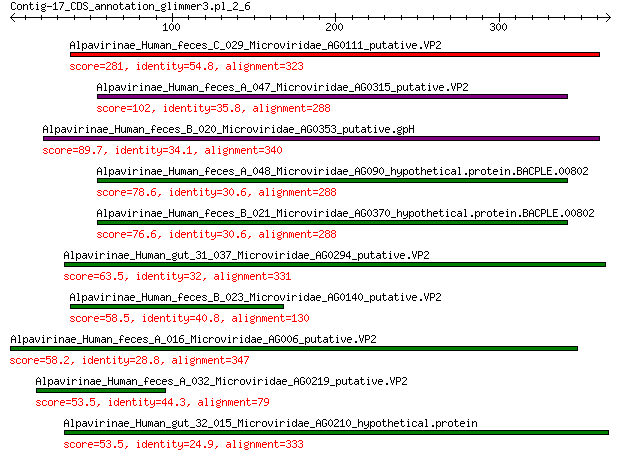
Alpavirinae_Human_feces_C_029_Microviridae_AG0111_putative.VP2 281 1e-93
Alpavirinae_Human_feces_A_047_Microviridae_AG0315_putative.VP2 102 2e-26
Alpavirinae_Human_feces_B_020_Microviridae_AG0353_putative.gpH 89.7 6e-22
Alpavirinae_Human_feces_A_048_Microviridae_AG090_hypothetical.p... 78.6 4e-18
Alpavirinae_Human_feces_B_021_Microviridae_AG0370_hypothetical.... 76.6 2e-17
Alpavirinae_Human_gut_31_037_Microviridae_AG0294_putative.VP2 63.5 5e-13
Alpavirinae_Human_feces_B_023_Microviridae_AG0140_putative.VP2 58.5 2e-11
Alpavirinae_Human_feces_A_016_Microviridae_AG006_putative.VP2 58.2 3e-11
Alpavirinae_Human_feces_A_032_Microviridae_AG0219_putative.VP2 53.5 8e-10
Alpavirinae_Human_gut_32_015_Microviridae_AG0210_hypothetical.p... 53.5 8e-10
> Alpavirinae_Human_feces_C_029_Microviridae_AG0111_putative.VP2
Length=329
Score = 281 bits (720), Expect = 1e-93, Method: Compositional matrix adjust.
Identities = 177/326 (54%), Positives = 237/326 (73%), Gaps = 5/326 (2%)
Query 38 MSAKDAKRQHQYDLEKMAVQHGYNIESQKLGQQFNKEMWDYTNYENQKKHLEAAGLNPal 97
MS+ A Q L+ M +Q+ YN + K QQ NK++WDYTNYENQK+H+E AGLNPAL
Sbjct 1 MSSGAANEQWGNQLKLMEIQNRYNEQMAKNNQQRNKDLWDYTNYENQKQHIENAGLNPAL 60
Query 98 lygmgggggataagaqglgagiasgH--EMGIKQQGRGMGLQTAAIASQIDLNNSQAEKN 155
+YGMGGGGG +A GAQG G + EMG+KQQ G+GLQ A+IASQ+DLN SQAEKN
Sbjct 61 MYGMGGGGGISANGAQGQGVTQPTDRSVEMGLKQQ--GLGLQLASIASQVDLNKSQAEKN 118
Query 156 RAEAKKTAGVDTNLQNANIEYVIAQTSNEKVKKGLIYANTRLLDAQEELQRTSVDYTKQK 215
+AEA+K +GVDT Q A I+ +IAQTSNEKVKKGLI R+ DA+EEL+R D+TK K
Sbjct 119 KAEAEKISGVDTRAQEATIDNLIAQTSNEKVKKGLILGQIRVADAEEELKRNMADWTKDK 178
Query 216 TDEVRWNIKLIEKEVDRLAKEINGLDLDNEYKKETLKNRITQTSLaiqqaiadiaLKGSQ 275
DE RWNIK ++K +D+L +EING+ LDNE K+ T+ N++ ++SL +Q +A+I LKGSQ
Sbjct 179 ADETRWNIKSLQKGIDKLIEEINGMKLDNELKERTIDNKVKESSLTLQNLMAEILLKGSQ 238
Query 276 GRLNDAEAKAIPEKILQMWAEIRTKQESVEVSKGQMENYAQDIANRLHLGEKGLDIQEQK 335
++N+ +AKAIP +ILQ W ++ + +++ + Q+E Y QD+ NR LG+KGLDI+EQK
Sbjct 239 RKVNEEQAKAIPAEILQGWEKLVKEGKALINQRDQIEAYVQDVINRYELGKKGLDIEEQK 298
Query 336 LIIDCITGLLNIAVQGGLTPMG-KVG 360
L+ D I G+L IA +G +G KVG
Sbjct 299 LVKDVILGMLEIASKGAGAALGAKVG 324
> Alpavirinae_Human_feces_A_047_Microviridae_AG0315_putative.VP2
Length=325
Score = 102 bits (254), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 103/290 (36%), Positives = 165/290 (57%), Gaps = 25/290 (9%)
Query 54 MAVQHGYNIESQKLGQQFNKEMWDYTNYENQKKHLEAAGLNPallygmgggggataagaq 113
M +Q+ YN + K QQ NK++WDYTNYENQK+H++ AGLNPAL+YGMGGGGG +A GAQ
Sbjct 1 MEIQNRYNEQMAKNNQQRNKDLWDYTNYENQKQHIKNAGLNPALMYGMGGGGGVSANGAQ 60
Query 114 glgagiasgHEMGIKQQGRGMGLQTAAIASQIDLNNSQAEKNRAEAKKTAGVDTNLQNAN 173
G G + + +K + +G+GLQ A+IASQ++LN S AEKN+ EA K AG DT +
Sbjct 61 GQGVTQPTDRSVEMKLKQQGLGLQLASIASQVELNKSLAEKNKVEADKIAGADTKVAEKQ 120
Query 174 IEYVIAQTSNEKVKKGLIYANTRLLDAQEELQRTSVDYTKQKTDEVRWNIKLIEKEV-DR 232
E + +Q+ K L + +L +AQE Q+T+ +Y + I+ EK+V +
Sbjct 121 AEMLESQSEFNKRITKLQDSIEKLTNAQE--QKTAAEY---------FYIQAQEKKVWEE 169
Query 233 LAKEINGLDLDNEYKKETLKNRITQTSLaiqqaiadiaLKG-SQGRLNDAEAKAIPEKIL 291
+ +++ D+ E K + Q ++ + ++ ++ +LN+ + + +I
Sbjct 170 VREQVVKADV-----AENTKEAMIQKAVLENFNLMQTGIESITRQKLNNEQINYLKGQIA 224
Query 292 QMWAEIRTKQESVEVSKGQMENYAQDIANRLHLGEKGLDIQEQKLIIDCI 341
WA + ++SV N + IAN L +G + LD ++++LI D I
Sbjct 225 IGWANVAIGEKSV-------SNESDRIANELMIGIRDLDRKDRELIKDWI 267
> Alpavirinae_Human_feces_B_020_Microviridae_AG0353_putative.gpH
Length=352
Score = 89.7 bits (221), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 116/361 (32%), Positives = 176/361 (49%), Gaps = 57/361 (16%)
Query 21 MIPGFAISTASNAVGG---AMSAKDAK-RQHQYDLEKMAVQHGYNIESQKLGQQFNKEMW 76
M+ G I+ A++A+G + A+DA+ R + E M Q+ N + + QQ K MW
Sbjct 1 MLGGI-IAAATSAIGSQIKGIEARDAEERAYTKQKELMDKQYALNDKMAEANQQRAKYMW 59
Query 77 DYTNYENQKKHLEAAGLNPallygmgggggataagaqglgagiasgHEMGIKQQGRGMGL 136
DYTN+ENQK+HL A L+P L YG G GG+T +G QG G G+ + +G Q + +GL
Sbjct 60 DYTNFENQKQHLLNANLSPGLFYGGSGAGGSTTSGGQGSGVGLGTETGVGYGIQEKALGL 119
Query 137 QTAAIASQIDLNNSQAEKNRAEAKKTAGVDTNLQNANIEYVIAQTSNEKVKKGLIYANTR 196
Q A++ASQ+ LN SQA KN AEAKK +GVDT L + T+
Sbjct 120 QLASMASQVALNQSQANKNNAEAKKISGVDTQLTE---------------------SQTK 158
Query 197 LLDAQEELQRTSVDYTKQKTDEVRWNIKLIEKEVDRLAKEINGLDLDN---EYKKETLKN 253
L A E L TK++ + + + L +E ++ +E + L N E K+T +
Sbjct 159 LNKAMENLTN-----TKEQREAADYFVAL--QEQSKVFEEARAMALQNDITEATKQTQID 211
Query 254 RITQ----TSLaiqqaiadiaLKGSQGRLNDAEAKAIPEKILQMWAEIRTKQESVEVSKG 309
+ Q SL + IA I LKG EA I ++I E TK+ S E +
Sbjct 212 TVVQNYYLNSLTAFEKIAGIELKGQ-------EAAYISKQIEWYSFEAITKRMSAEAMQS 264
Query 310 QMENYAQDIANRLHLGEKGLDIQEQKL----IIDCITGLLNIAVQGG------LTPMGKV 359
++ A+ + N + K LD +++++ I + + L +A G P+ KV
Sbjct 265 MAKSAAERVKNDFEIAGKKLDQEQERILQNWIFESVKSLCTVAETTGDIISMFRKPIQKV 324
Query 360 G 360
G
Sbjct 325 G 325
> Alpavirinae_Human_feces_A_048_Microviridae_AG090_hypothetical.protein.BACPLE.00802
Length=333
Score = 78.6 bits (192), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 88/288 (31%), Positives = 136/288 (47%), Gaps = 22/288 (8%)
Query 54 MAVQHGYNIESQKLGQQFNKEMWDYTNYENQKKHLEAAGLNPallygmgggggataagaq 113
M Q N E K WDYTNYEN KHL+ AGLNPAL Y GG GGAT G
Sbjct 10 MNKQAELNKEQADYSTDLAKNYWDYTNYENSVKHLKEAGLNPALFYAKGGQGGATGGGQA 69
Query 114 glgagiasgHEMGIKQQGRGMGLQTAAIASQIDLNNSQAEKNRAEAKKTAGVDTNLQNAN 173
+ M + Q +GMG Q + SQ++LN + A+K AEA+K AG DT +
Sbjct 70 QGVGLPPTTPTMA-RIQAQGMGAQLQNVLSQVELNKATAKKTEAEAEKIAGADTKVAERE 128
Query 174 IEYVIAQTSNEKVKKGLIYANTRLLDAQEELQRTSVDYTKQKTDEVRWNIKLIEKEVDRL 233
E + +Q+ + L + +L +AQE Q+T+ +Y + E ++
Sbjct 129 AEMLESQSEFNRRITRLQDSIEKLTNAQE--QKTAAEYFYTQAQE------------KKV 174
Query 234 AKEINGLDLDNEYKKETLKNRITQTSLaiqqaiadiaLKGSQGRLNDAEAKAIPEKILQM 293
+E+ + ++ +ET + I +T L + ++ +LN + + ++
Sbjct 175 WEEVREQIVKSDVAEETKEAMIRKTGLENFNLMQAGIESITRQKLNSEQINYLRGQLAIG 234
Query 294 WAEIRTKQESVEVSKGQMENYAQDIANRLHLGEKGLDIQEQKLIIDCI 341
WA + ++SV N A IAN L +G K LD ++++LI D I
Sbjct 235 WANVAIGEKSV-------SNEADRIANELMMGMKDLDRKDRELIKDWI 275
> Alpavirinae_Human_feces_B_021_Microviridae_AG0370_hypothetical.protein.BACPLE.00802
Length=333
Score = 76.6 bits (187), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 88/288 (31%), Positives = 135/288 (47%), Gaps = 22/288 (8%)
Query 54 MAVQHGYNIESQKLGQQFNKEMWDYTNYENQKKHLEAAGLNPallygmgggggataagaq 113
M Q N E K WDYTNYEN KHL+ AGLNPAL Y GG GGAT G
Sbjct 10 MNKQAELNKEQADYSTDLAKNYWDYTNYENSVKHLKEAGLNPALFYAKGGQGGATGGGQA 69
Query 114 glgagiasgHEMGIKQQGRGMGLQTAAIASQIDLNNSQAEKNRAEAKKTAGVDTNLQNAN 173
++ M + Q +GMG Q + SQ++LN + A+K AEA+K AG DT +
Sbjct 70 QGVGLPSTTPTMA-RIQAQGMGAQLQNVLSQVELNKATAKKTEAEAEKIAGADTKVAERE 128
Query 174 IEYVIAQTSNEKVKKGLIYANTRLLDAQEELQRTSVDYTKQKTDEVRWNIKLIEKEVDRL 233
E + +Q+ K L + +L AQE Q+T+ +Y + E ++
Sbjct 129 AEMLESQSEFNKRVTKLQDSIEKLNKAQE--QKTAAEYFYTQAQE------------KKV 174
Query 234 AKEINGLDLDNEYKKETLKNRITQTSLaiqqaiadiaLKGSQGRLNDAEAKAIPEKILQM 293
+E+ + ++ +ET + I + L + ++ +LN + + ++
Sbjct 175 WEEVREQIVKSDVAEETKEAMIERAGLENFNLMQAGIESITRQKLNSEQINYLKGQLAIG 234
Query 294 WAEIRTKQESVEVSKGQMENYAQDIANRLHLGEKGLDIQEQKLIIDCI 341
WA + ++SV N A IAN L +G K LD ++++LI D I
Sbjct 235 WANVAIGEKSV-------SNEADRIANELMMGMKDLDRKDRELIKDWI 275
> Alpavirinae_Human_gut_31_037_Microviridae_AG0294_putative.VP2
Length=360
Score = 63.5 bits (153), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 106/364 (29%), Positives = 163/364 (45%), Gaps = 71/364 (20%)
Query 34 VGGAMSAKDAKRQHQYDLEKMAVQHGYNIESQKLGQQF---------------------- 71
+GGA+S + R ++ L+ M QH YN E ++ Q +
Sbjct 2 IGGAISDR---RNYKNQLKLMGQQHQYNKEMGEINQNYAKEMAMINNQYALGMAKESHNM 58
Query 72 NKEMWDYTNYENQKKHLEAAGLNPallygmgggggataagaqglgagiasgHEMG----- 126
NK+MW+YTNYENQ H++AAGLNPALLYG GGGGGATA G + G G
Sbjct 59 NKDMWNYTNYENQVAHMKAAGLNPALLYGNGGGGGATATGGTAIPGQGTPGSAPGGAGPQ 118
Query 127 -IKQQ---GRGMGLQTAAIASQIDLNNSQAEKNRAEAKKTAGVDTNLQNANIEYVIAQTS 182
IK Q GMG+Q + +Q + A K AEA KTAGVDT L A+T+
Sbjct 119 AIKSQIIESTGMGIQLGLMNAQKRNLEADAAKKEAEATKTAGVDTEL---------AKTA 169
Query 183 NEKVKKGLIYANTRLLDAQEELQRTSVDYTKQKTDEVRWNIKLIEKEVDRLAKEINGLDL 242
+L ++ T T+E+ K+ L ++
Sbjct 170 -------------------AKLNEAKIENTNMSTEEIAAKAKMWGDTSTVLWQQARKYAS 210
Query 243 DNEYKKETLKNRITQTSLaiqqaiadiaLKGSQGRLNDAEAKAIPEKILQMWAEIRTKQE 302
+ +Y ++T+ RI + ++ + ++ + +A+ KAI E I W T +
Sbjct 211 EADYNEKTMDTRIEKVGYDTMGSLLENMETIAKTQFTEAQTKAITENIAIAWYNAGTNRM 270
Query 303 SVEVSKGQMENYAQDIANRLHLGEKGLDIQEQKLIIDCITGLLN--IAVQGGLTPMGKVG 360
+ + A +AN L LDI+E++L+ D I ++ +A+ G+T M KV
Sbjct 271 NATTA-------ADHVANELFKTMGELDIKERQLLKDWIYQGVHAGVALIEGVTDMVKVK 323
Query 361 SIAK 364
++ K
Sbjct 324 ALIK 327
> Alpavirinae_Human_feces_B_023_Microviridae_AG0140_putative.VP2
Length=295
Score = 58.5 bits (140), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 53/132 (40%), Positives = 71/132 (54%), Gaps = 9/132 (7%)
Query 38 MSAKDAKRQHQYDLEK--MAVQHGYNIESQKLGQQFNKEMWDYTNYENQKKHLEAAGLNP 95
M++++ Q Q+ LEK MA+Q YN E QQ +MW+ TNYE+Q +H++AAGLNP
Sbjct 1 MNSQEQAMQDQWKLEKEKMALQAKYNKEQADYSQQLALDMWNATNYESQVEHMKAAGLNP 60
Query 96 allygmgggggataagaqglgagiasgHEMGIKQQGRGMGLQTAAIASQIDLNNSQAEKN 155
ALLY GG GG+T+ + Q GMGLQ IA ++ K
Sbjct 61 ALLYSKGGAGGSTSGAGTAAPVSDGTT-------QAVGMGLQAKQIAISQAQQMAETAKT 113
Query 156 RAEAKKTAGVDT 167
AE K +GVDT
Sbjct 114 VAETAKISGVDT 125
> Alpavirinae_Human_feces_A_016_Microviridae_AG006_putative.VP2
Length=367
Score = 58.2 bits (139), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 100/370 (27%), Positives = 170/370 (46%), Gaps = 52/370 (14%)
Query 1 MSFWSKIGDAVKSIGKG--IVNMIPGFAISTASNAVGGAM----------SAKD----AK 44
MSF I A++ KG I N+IPG + GG S KD +
Sbjct 1 MSFGLGISGALEGASKGTSIGNIIPGLGTTAGGIIGGGLGLLGGLFGGGSSTKDQEKLME 60
Query 45 RQHQYDLEKMAVQHGYNIESQKLGQQFNKEMWDYTNYENQKKHLEAAGLNPallygmggg 104
+ +Y+ E M +Q+ Y + Q+ N EMW+ TN+ Q++H+E AGL+ L GGG
Sbjct 61 KAWEYEKEGMGMQYQYGQAAANEAQRRNMEMWNQTNFGAQRQHMEDAGLS-VGLMYGGGG 119
Query 105 ggataagaqglgagiasgHEMGIKQQGRGMGLQTAAIASQIDLNNSQAEKNRAEAKKTAG 164
GA + G Q + + +G+ Q + + Q AI SQ LN ++A K AEAKKT G
Sbjct 120 QGAVSQGGQATQPSGPTSNPVGMALQYKQIEQQNEAIKSQTMLNQAEAAKALAEAKKTGG 179
Query 165 VDTNLQNANIEYVIAQTSNEKVKKGLIYANTRLLDAQEELQRTSVDYTKQKTDEVRWNIK 224
VDT + I++ ++++ R+ +++E++ +++ E + N K
Sbjct 180 VDTKKTESEIKW-------QEIE-------NRIQESREQIASSNI-------IEAKANAK 218
Query 225 LIEKEVDRLAKEINGLDLDNEYKKETLKNRITQTSLaiqqaiadiaLKGSQGRLNDAEAK 284
+ +E L+ EY +T + RI + + + + L +A+A
Sbjct 219 -------KTVEEFKQAMLNTEYLDKTQQQRIQMVTDQLSLIQKQGLKEEAVIDLTNAQAS 271
Query 285 AIPEKILQMWAEIRTKQESVEVSKGQMENYAQDIANRLHLGEKGLDIQEQKLI------- 337
+ ++I +W + TK+ S + K Q + IA LG+ L ++EQK +
Sbjct 272 KVRKEIDILWYDAITKRTSADALKKQADTAVDKIAKEYELGKGKLSLEEQKNLREWIYGG 331
Query 338 IDCITGLLNI 347
ID ITG++ +
Sbjct 332 IDQITGIVEV 341
> Alpavirinae_Human_feces_A_032_Microviridae_AG0219_putative.VP2
Length=355
Score = 53.5 bits (127), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 35/103 (34%), Positives = 50/103 (49%), Gaps = 26/103 (25%)
Query 17 GIVNMI----PGFAISTASNAVGGAMSAKDAKRQHQYDLEKMAVQHG------------- 59
G++N I G A G S+KD R HQ L+K +++G
Sbjct 2 GLLNFINKGGGGLITGLAGTIAGAVSSSKD--RAHQEKLQKQQMEYGREMYALQSADEDR 59
Query 60 -------YNIESQKLGQQFNKEMWDYTNYENQKKHLEAAGLNP 95
+N E+ + Q++ KEM+DYT YENQ K ++AAGLNP
Sbjct 60 RMEQQNQWNKEAAEQSQEYAKEMFDYTGYENQVKQMKAAGLNP 102
> Alpavirinae_Human_gut_32_015_Microviridae_AG0210_hypothetical.protein
Length=341
Score = 53.5 bits (127), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 83/334 (25%), Positives = 152/334 (46%), Gaps = 39/334 (12%)
Query 34 VGGAMSAKDA-KRQHQYDLEKMAVQHGYNIESQKLGQQFNKEMWDYTNYENQKKHLEAAG 92
+GG ++ K+ K+Q +++ E M +Q+ YN + EMW+ T YE Q + +E AG
Sbjct 38 IGGWITGKNKEKKQREHEKEMMGLQYQYNEAAANNNMTRALEMWEKTGYEAQGQQIENAG 97
Query 93 LNPallygmgggggataagaqglgagiasgHEMGIKQQGRGMGLQTAAIASQIDLNNSQA 152
LN AL+YG GG + + + Q MGLQ A+ +Q+ +
Sbjct 98 LNKALMYGGGGASATSQSQGNSGVNNTGT--------QAVAMGLQARAMEAQVSNTEADT 149
Query 153 EKNRAEAKKTAGVDTNLQNANIEYVIAQTSNEKVKKGLIYANTRLLDAQEELQRTSVDYT 212
A+A K AG IE I QT N + D ++ L+ +V+
Sbjct 150 ALKIAQAAKEAG---EASKKPIELKIEQT------------NKEITDLEKSLKAQNVEIG 194
Query 213 KQKTDEVRWNIKLIEKEVDRLAKEINGLDLDNEYKKETLKNRITQTSLaiqqaiadiaLK 272
+E+ + + +K ++ E+N ++ E KKET I T + IAL
Sbjct 195 ---ANEMVRSTAIAQKAME----ELNQAMMETEIKKETKDAIIKSTIKNVTNLEVQIALG 247
Query 273 GSQGRLNDAEAKAIPEKILQMWAEIRTKQESVEVSKGQMENYAQDIANRLHLGEKGLDIQ 332
++ + + +AI ++ + ++ T+Q + E +K + + + + + + LD+Q
Sbjct 248 IAKTKETNKNIEAIGGQLEALKKDVITRQITAEAAKENAKTLGERLIKEMEVKGQELDLQ 307
Query 333 EQKLIIDCITGLLNIAVQGGLTPMGKVGSIAKGG 366
E+++I+D A++GG+ + K G + KGG
Sbjct 308 EKRMILD--------AIKGGVESILKFGFLKKGG 333
Lambda K H a alpha
0.311 0.128 0.351 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 31835727