bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-13_CDS_annotation_glimmer3.pl_2_2
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
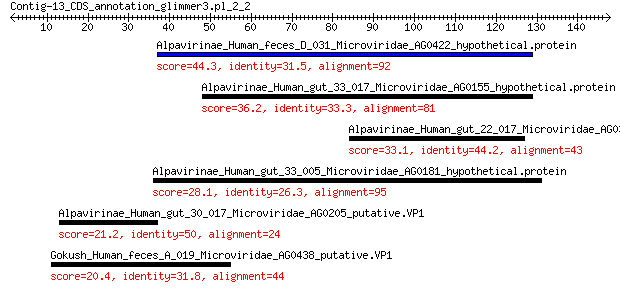
Alpavirinae_Human_feces_D_031_Microviridae_AG0422_hypothetical.... 44.3 3e-08
Alpavirinae_Human_gut_33_017_Microviridae_AG0155_hypothetical.p... 36.2 4e-05
Alpavirinae_Human_gut_22_017_Microviridae_AG0398_hypothetical.p... 33.1 1e-04
Alpavirinae_Human_gut_33_005_Microviridae_AG0181_hypothetical.p... 28.1 0.020
Alpavirinae_Human_gut_30_017_Microviridae_AG0205_putative.VP1 21.2 5.9
Gokush_Human_feces_A_019_Microviridae_AG0438_putative.VP1 20.4 8.4
> Alpavirinae_Human_feces_D_031_Microviridae_AG0422_hypothetical.protein
Length=150
Score = 44.3 bits (103), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 29/96 (30%), Positives = 48/96 (50%), Gaps = 4/96 (4%)
Query 37 YDETSDGDLIQCDMTQILLNQEKYRRLLGDMNIQNILAQMHPTQSTVMDGMTDEDRFACV 96
Y + DG + +L+N E+ R +G+ + N++ + P +S + TDE F +
Sbjct 38 YVKDDDGVIRYVSDVNLLMNAERLRNQIGEESYLNLIRGIQPKKSPYDNKYTDEQLFTAI 97
Query 97 ISRHCQTMSERQAVLQQLASE----KSELTAYAEAM 128
SR QT SE A ++ L S +SEL A E++
Sbjct 98 KSRFIQTPSEVLAWIESLGSAGDSIRSELDALTESV 133
> Alpavirinae_Human_gut_33_017_Microviridae_AG0155_hypothetical.protein
Length=171
Score = 36.2 bits (82), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 27/82 (33%), Positives = 43/82 (52%), Gaps = 8/82 (10%)
Query 48 CDMTQILLNQEKYRRLLGDMNIQNILAQMHPTQ-STVMDGMTDEDRFACVISRHCQTMSE 106
D+ IL N++ R D+ + ++ P+Q +MD M+DED A V SR+ Q+ SE
Sbjct 66 SDIRLILHNKDLASRAGVDVASKFGQSKQSPSQIQQIMDTMSDEDLLATVRSRYIQSPSE 125
Query 107 RQAVLQQLASEKSELTAYAEAM 128
+ + EL+AYAE +
Sbjct 126 -------ILAWSKELSAYAENL 140
> Alpavirinae_Human_gut_22_017_Microviridae_AG0398_hypothetical.protein
Length=69
Score = 33.1 bits (74), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 19/43 (44%), Positives = 24/43 (56%), Gaps = 7/43 (16%)
Query 84 MDGMTDEDRFACVISRHCQTMSERQAVLQQLASEKSELTAYAE 126
MD M+D+D A V SRH Q SE + + EL+AYAE
Sbjct 1 MDTMSDDDLLATVRSRHIQAPSE-------IIAWSKELSAYAE 36
> Alpavirinae_Human_gut_33_005_Microviridae_AG0181_hypothetical.protein
Length=205
Score = 28.1 bits (61), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 25/102 (25%), Positives = 46/102 (45%), Gaps = 11/102 (11%)
Query 36 MYDETSDGDLIQCDMTQILLNQEKYRRLLGDMN---IQNILAQMHPTQSTVMDGMTDEDR 92
++D +DG + C IL Q + + +MN ++ + + P S D+
Sbjct 90 LFDHNADGSVTFCSDYGILFGQ----KAIDNMNQVQLRRYMNSLVPRSSNYTRNYNDDFL 145
Query 93 FACVISRHCQTMSERQAVLQQLASE----KSELTAYAEAMLA 130
R+ Q+ +E + L L SE +S+L AYA+++ A
Sbjct 146 LDYCKDRNIQSATEMASWLDHLLSEGQSLESDLQAYADSLSA 187
> Alpavirinae_Human_gut_30_017_Microviridae_AG0205_putative.VP1
Length=647
Score = 21.2 bits (43), Expect = 5.9, Method: Composition-based stats.
Identities = 12/27 (44%), Positives = 14/27 (52%), Gaps = 3/27 (11%)
Query 13 KDDYVPEL-VENHPCYQE--SVYDSVM 36
K D V + V PCY E S YD V+
Sbjct 535 KSDAVSSVSVAKEPCYNEFRSSYDEVL 561
> Gokush_Human_feces_A_019_Microviridae_AG0438_putative.VP1
Length=569
Score = 20.4 bits (41), Expect = 8.4, Method: Composition-based stats.
Identities = 14/53 (26%), Positives = 21/53 (40%), Gaps = 9/53 (17%)
Query 11 FVKDDYVPELVENHPCYQESVYD---------SVMYDETSDGDLIQCDMTQIL 54
F Y EL Y+E +Y +V E GD I +M+Q++
Sbjct 326 FQVQKYFEELARGGSRYREQIYSLFRTRISDKTVQIPEYLGGDRIMINMSQVV 378
Lambda K H a alpha
0.317 0.129 0.368 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 9663476